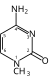
Hachimoji DNA

Hachimoji DNA (from Japanese 八文字 hachimoji, "eight letters") is a synthetic nucleic acid analog that uses four synthetic nucleotides in addition to the four present in the natural nucleic acids, DNA and RNA. This leads to four allowed base pairs: two unnatural base pairs formed by the synthetic nucleobases in addition to the two normal pairs. Hachimoji bases have been demonstrated in both DNA and RNA analogs, using deoxyribose and ribose respectively as the backbone sugar.
Benefits of such a nucleic acid system may include an enhanced ability to store data, as well as insights into what may be possible in the search for extraterrestrial life.
The hachimoji DNA system produced one type of catalytic RNA (ribozyme or aptamer) in vitro.
Description
Natural DNA is a molecule carrying the genetic instructions used in the growth, development, functioning, and reproduction of all known living organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids; alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life. DNA is a polynucleotide as it is composed of simpler monomeric units called nucleotides; when double-stranded, the two chains coil around each other to form a double helix.
In natural DNA, each nucleotide is composed of one of four nucleobases (cytosine [C], guanine [G], adenine [A] or thymine [T]), a sugar called deoxyribose, and a phosphate group. The nucleotides are joined to one another in a chain by covalent bonds between the sugar of one nucleotide and the phosphate of the next, resulting in an alternating sugar-phosphate backbone. The nitrogenous bases of the two separate polynucleotide strands are bound to each other with hydrogen bonds, according to base pairing rules (A with T and C with G), to make double-stranded DNA.
Hachimoji DNA is similar to natural DNA but differs in the number, and type, of nucleobases. Unnatural nucleobases, more hydrophobic than natural bases, are used in successful hachimoji DNA. Such a DNA always formed the standard double helix, no matter what sequence of bases were used. An enzyme (T7 polymerase) was adapted by the researchers to be used in vitro to transcribe hachimoji DNA into hachimoji RNA, which, in turn, produced chemical activity in the form of a glowing green fluorophore.
New base pairs
DNA and RNA are naturally composed of four nucleotide bases that form hydrogen bonds in order to pair. Hachimoji DNA uses an additional four synthetic nucleotides to form four types of base pairs, two of which are unnatural: P binds with Z and B binds with S (dS in DNA, rS in RNA).
Base Name Formula SMILES Structure ChemSpider PubChem P 2-Aminoimidazo[1,2a][1,3,5]triazin-4(1H)-one 2-amino-8-(1′-b-D-2′-deoxyribofuranosyl)-imidazo-[1,2a]-1,3,5-triazin-[8H]-4-one
C5H5N5O C1=CN2C(=O)NC(=NC2=N1)N 10205066 135600909 Z 6-Amino-5-nitropyridin-2-one 6-amino-3-(1′-b-D-2′-deoxyribofuranosyl)-5-nitro-1H-pyridin-2-one
C5H5N3O3 C1=CC(=O)NC(=C1[N+](=O)[O-])N 9357814 11182729 B Isoguanine 6-amino-9[(1′-b-D-2′-deoxyribofuranosyl)-4-hydroxy-5-(hydroxymethyl)-oxolan-2-yl]-1H-purin-2-one
C5H5N5O C1=NC2=NC(=O)NC(=C2N1)N 69351 76900 S rS Isocytosine C4H5N3O C1=CN=C(NC1=O)N 60309 66950 dS 1-Methylcytosine 3-methyl-6-amino-5-(1′-b-D-2′-deoxyribofuranosyl)-pyrimidin-2-one
C5H7N3O CN1C=CC(=NC1=O)N 71474 79143
The natural bases are in the upper row; the unnatural, synthetic bases are in the lower row.
Hydrogen bonds are dashed green lines, with acceptor atoms in red.
Background
Earlier, the research group responsible for the hachimoji DNA system, headed by Harvard University chemist Steven Benner, had studied a synthetic DNA analog system, named Artificially Expanded Genetic Information System (AEGIS), that used twelve different nucleotides, including the four found in DNA.
Biology
Scripps Research chemist Floyd Romesberg, noted for creating the first Unnatural Base Pair (UBP), and expanding the genetic alphabet of four letters to six in 2012, stated that the invention of the hachimoji DNA system is an example of the fact that the natural bases (G, C, A and T) "are not unique". Creating new life forms may be possible, at least theoretically, with the new DNA system. For now, however, the hachimoji DNA system is not self-sustaining; the system needs a steady supply of unique building blocks and proteins found only in the laboratory. As a result, "Hachimoji DNA can go nowhere if it escapes the laboratory."
Applications
NASA funded this research to "expand[s] the scope of the structures that we might encounter as we search for life in the cosmos". According to Lori Glaze of the Planetary Science Division of NASA, "Life detection is an increasingly important goal of NASA's planetary science missions, and this new work [with hachimoji DNA] will help us to develop effective instruments and experiments that will expand the scope of what we look for." Research team leader Steven Benner notes, "By carefully analyzing the roles of shape, size and structure in hachimoji DNA, this work expands our understanding of the types of molecules that might store information in extraterrestrial life on alien worlds."
According to researchers, hachimoji DNA could also be used "to develop clean diagnostics for human diseases, in DNA digital data storage, DNA barcoding, self-assembling nanostructures, and to make proteins with unusual amino acids. Parts of this hachimoji DNA are already being commercially produced by Firebird Biomolecular Sciences LLC".
See also
Further reading
- Bains W (2004). "Many chemistries could be used to build living systems" (PDF). Astrobiology. 4 (2): 137–67. Bibcode:2004AsBio...4..137B. doi:10.1089/153110704323175124. PMID 15253836. Hypothesis paper.
External links
- Astronomy FAQ - Why do we assume that other beings must be based on carbon? Why couldn't organisms be based on other substances?
- Film clip encoded into DNA using CRISPR: video (01:39) on YouTube
| Key components | |
|---|---|
| Fields | |
| Archaeogenetics of | |
| Related topics | |
| Lists | |
| Introduction to genetics |
|||||||
|---|---|---|---|---|---|---|---|
| Transcription |
|
||||||
| Translation |
|
||||||
| Regulation | |||||||
| Influential people | |||||||
|
Types of nucleic acids
| |||||||
|---|---|---|---|---|---|---|---|
| Constituents | |||||||
|
Ribonucleic acids |
|
||||||
| Deoxyribonucleic acids |
|||||||
| Analogues | |||||||
| Cloning vectors | |||||||