Iron-sulfur protein
Iron–sulfur proteins are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulnerable to attack by biogenic nitric oxide, forming dinitrosyl iron complexes. In most Fe–S proteins, the terminal ligands on Fe are thiolate, but exceptions exist.
The prevalence of these proteins on the metabolic pathways of most organisms leads some scientists to theorize that iron–sulfur compounds had a significant role in the origin of life in the iron–sulfur world theory.
Structural motifs
In almost all Fe–S proteins, the Fe centers are tetrahedral and the terminal ligands are thiolato sulfur centers from cysteinyl residues. The sulfide groups are either two- or three-coordinated. Three distinct kinds of Fe–S clusters with these features are most common.
Structure-Function Principles
To serve their various biological roles, iron-sulfur proteins effect rapid electron transfers and span the whole range of physiological redox potentials from -600 mV to +460 mV.
Iron-sulfur proteins are involved in various biological electron transport processes, such as photosynthesis and cellular respiration, which require rapid electron transfer to sustain the energy or biochemical needs of the organism.
Fe3+-SR bonds have unusually high covalency which is expected. When comparing the covalency of Fe3+ with the covalency of Fe2+, Fe3+ has almost double the covalency of Fe2+ (20% to 38.4%). Fe3+ is also much more stabilized than Fe2+. Hard ions like Fe3+ normally have low covalency because of the energy mismatch of the metal Lowest Unoccupied Molecular Orbital with the ligand Highest Occupied Molecular Orbital.
There is HO-H—S-Cys H-bonding from external H2O’s positioned by the protein close to the active site and this H-bonding decreases the lone pair electron donation from the Cys-S donor to the Fe3+/2+. Using lyophilization to remove these external H2O’s results in increased Fe-S covalency, which means that the H2O’s are decreasing the covalency because HOH-S Hydrogen-bonding pulls the sulfur electrons. Since covalency stabilizes Fe3+ more than Fe2+, therefore Fe3+ is more destabilized by the HOH-S hydrogen-bonding.
The Fe3+ 3d orbital energies follow the “inverted” bonding scheme which fortuitously has the Fe3+ d-orbitals closely matched in energy with the sulfur 3p orbitals which gives high covalency in the resulting bonding molecular orbital. This high covalency lowers the inner sphere reorganization energy and ultimately contributes to a rapid electron transfer.
2Fe–2S clusters
The simplest polymetallic system, the [Fe2S2] cluster, is constituted by two iron ions bridged by two sulfide ions and coordinated by four cysteinyl ligands (in Fe2S2ferredoxins) or by two cysteines and two histidines (in Rieske proteins). The oxidized proteins contain two Fe3+ ions, whereas the reduced proteins contain one Fe3+ and one Fe2+ ion. These species exist in two oxidation states, (FeIII)2 and FeIIIFeII. CDGSH iron sulfur domain is also associated with 2Fe-2S clusters.
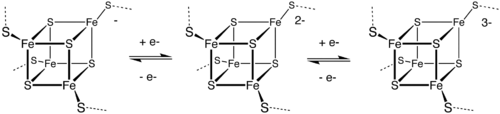
4Fe–4S clusters
A common motif features a four iron ions and four sulfide ions placed at the vertices of a cubane-type cluster. The Fe centers are typically further coordinated by cysteinyl ligands. The [Fe4S4] electron-transfer proteins ([Fe4S4] ferredoxins) may be further subdivided into low-potential (bacterial-type) and high-potential (HiPIP) ferredoxins. Low- and high-potential ferredoxins are related by the following redox scheme:
In HiPIP, the cluster shuttles between [2Fe3+, 2Fe2+] (Fe4S42+) and [3Fe3+, Fe2+] (Fe4S43+). The potentials for this redox couple range from 0.4 to 0.1 V. In the bacterial ferredoxins, the pair of oxidation states are [Fe3+, 3Fe2+] (Fe4S4+) and [2Fe3+, 2Fe2+] (Fe4S42+). The potentials for this redox couple range from −0.3 to −0.7 V. The two families of 4Fe–4S clusters share the Fe4S42+ oxidation state. The difference in the redox couples is attributed to the degree of hydrogen bonding, which strongly modifies the basicity of the cysteinyl thiolate ligands. A further redox couple, which is still more reducing than the bacterial ferredoxins is implicated in the nitrogenase.
Some 4Fe–4S clusters bind substrates and are thus classified as enzyme cofactors. In aconitase, the Fe–S cluster binds aconitate at the one Fe centre that lacks a thiolate ligand. The cluster does not undergo redox, but serves as a Lewis acid catalyst to convert citrate to isocitrate. In radical SAM enzymes, the cluster binds and reduces S-adenosylmethionine to generate a radical, which is involved in many biosyntheses.
The second cubane shown here with mixed valence pairs (2 Fe3+ and 2 Fe2+), has a greater stability from covalent communication and strong covalent delocalization of the “extra” electron from the reduced Fe2+ that results in full ferromagnetic coupling.
3Fe–4S clusters
Proteins are also known to contain [Fe3S4] centres, which feature one iron less than the more common [Fe4S4] cores. Three sulfide ions bridge two iron ions each, while the fourth sulfide bridges three iron ions. Their formal oxidation states may vary from [Fe3S4]+ (all-Fe3+ form) to [Fe3S4]2− (all-Fe2+ form). In a number of iron–sulfur proteins, the [Fe4S4] cluster can be reversibly converted by oxidation and loss of one iron ion to a [Fe3S4] cluster. E.g., the inactive form of aconitase possesses an [Fe3S4] and is activated by addition of Fe2+ and reductant.
Other Fe–S clusters
More complex polymetallic systems are common. Examples include both the 8Fe and the 7Fe clusters in nitrogenase. Carbon monoxide dehydrogenase and the [FeFe]-hydrogenase also feature unusual Fe–S clusters. A special 6 cysteine-coordinated [Fe4S3] cluster was found in oxygen-tolerant membrane-bound [NiFe] hydrogenases.
Biosynthesis
The biosynthesis of the Fe–S clusters has been well studied. The biogenesis of iron sulfur clusters has been studied most extensively in the bacteria E. coli and A. vinelandii and yeast S. cerevisiae. At least three different biosynthetic systems have been identified so far, namely nif, suf, and isc systems, which were first identified in bacteria. The nif system is responsible for the clusters in the enzyme nitrogenase. The suf and isc systems are more general.
The yeast isc system is the best described. Several proteins constitute the biosynthetic machinery via the isc pathway. The process occurs in two major steps: (1) the Fe/S cluster is assembled on a scaffold protein followed by (2) transfer of the preformed cluster to the recipient proteins. The first step of this process occurs in the cytoplasm of prokaryotic organisms or in the mitochondria of eukaryotic organisms. In the higher organisms the clusters are therefore transported out of the mitochondrion to be incorporated into the extramitochondrial enzymes. These organisms also possess a set of proteins involved in the Fe/S clusters transport and incorporation processes that are not homologous to proteins found in prokaryotic systems.
Synthetic analogues
Synthetic analogues of the naturally occurring Fe–S clusters were first reported by Holm and coworkers. Treatment of iron salts with a mixture of thiolates and sulfide affords derivatives such as (Et4N)2Fe4S4(SCH2Ph)4].
See also
- Sticht, Heinrich; Rösch, Paul (1998-09-01). "The structure of iron–sulfur proteins". Progress in Biophysics and Molecular Biology. 70 (2): 95–136. doi:10.1016/S0079-6107(98)00027-3. ISSN 0079-6107. PMID 9785959.
Further reading
- Beinert, H. (2000). "Iron-sulfur proteins: ancient structures, still full of surprises". J. Biol. Inorg. Chem. 5 (1): 2–15. doi:10.1007/s007750050002. PMID 10766431. S2CID 20714007.
- Beinert, H.; Kiley, P.J. (1999). "Fe-S proteins in sensing and regulatory functions". Curr. Opin. Chem. Biol. 3 (2): 152–157. doi:10.1016/S1367-5931(99)80027-1. PMID 10226040.
- Johnson, M.K. (1998). "Iron-sulfur proteins: new roles for old clusters". Curr. Opin. Chem. Biol. 2 (2): 173–181. doi:10.1016/S1367-5931(98)80058-6. PMID 9667933.
- Nomenclature Committee of the International Union of Biochemistry (NC-IUB) (1979). "Nomenclature of iron-sulfur proteins. Recommendations 1978". Eur. J. Biochem. 93 (3): 427–430. doi:10.1111/j.1432-1033.1979.tb12839.x. PMID 421685.
-
Noodleman, L., Lovell, T., Liu, T., Himo, F. and Torres, R.A. (2002). "Insights into properties and energetics of iron-sulfur proteins from simple clusters to nitrogenase". Curr. Opin. Chem. Biol. 6 (2): 259–273. doi:10.1016/S1367-5931(02)00309-5. PMID 12039013.
{{cite journal}}: CS1 maint: multiple names: authors list (link) -
Spiro, T.G., Ed. (1982). Iron-sulfur proteins. New York: Wiley. ISBN 0-471-07738-0.
{{cite book}}: CS1 maint: multiple names: authors list (link)
External links
- Iron-Sulfur+Proteins at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Examples of iron-sulfur clusters
| heme | |
|---|---|
| nonheme | |
| Fe(-II) | |||
|---|---|---|---|
| Fe(0) | |||
| Fe(I) |
|
||
| Fe(0,II) | |||
| Fe(II) |
|
||
| Fe(0,III) | |||
| Fe(II,III) | |||
| Fe(III) |
|
||
| Fe(VI) | |||
| Purported | |||
|
Sulfur compounds
| |||||
|---|---|---|---|---|---|
|
Sulfides and disulfides |
|
||||
| Sulfur halides | |||||
| Sulfur oxides and oxyhalides |
|
||||
| Thiocyanates | |||||
| Organic compounds | |||||