Protein family
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be confused with family as it is used in taxonomy.
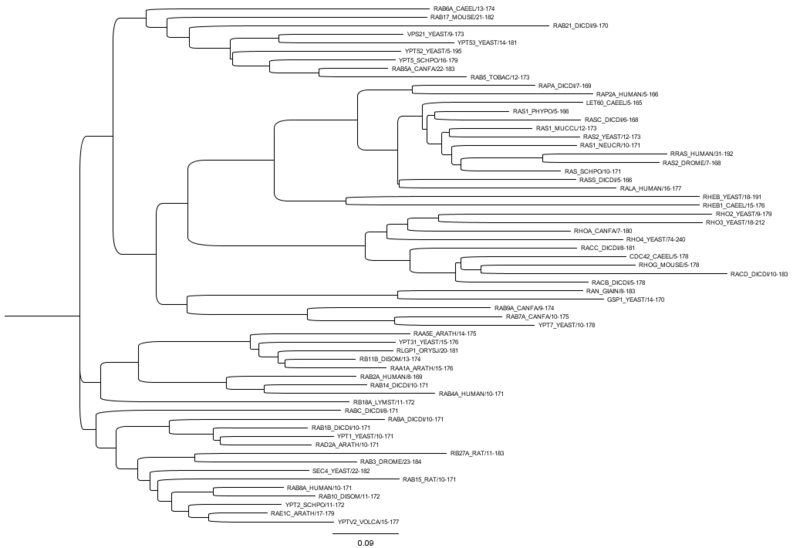
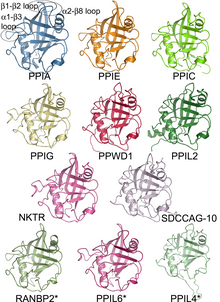
Proteins in a family descend from a common ancestor and typically have similar three-dimensional structures, functions, and significant sequence similarity. The most important of these is sequence similarity (usually amino-acid sequence), since it is the strictest indicator of homology and therefore the clearest indicator of common ancestry. A fairly well developed framework exists for evaluating the significance of similarity between a group of sequences using sequence alignment methods. Proteins that do not share a common ancestor are very unlikely to show statistically significant sequence similarity, making sequence alignment a powerful tool for identifying the members of protein families. Families are sometimes grouped together into larger clades called superfamilies based on structural and mechanistic similarity, even if no identifiable sequence homology is seen.
Currently, over 60,000 protein families have been defined, although ambiguity in the definition of "protein family" leads different researchers to highly varying numbers.
Terminology and usage
As with many biological terms, the use of protein family is somewhat context dependent; it may indicate large groups of proteins with the lowest possible level of detectable sequence similarity, or very narrow groups of proteins with almost identical sequence, function, and three-dimensional structure, or any kind of group in between. To distinguish between these situations, the term protein superfamily is often used for distantly related proteins whose relatedness is not detectable by sequence similarity, but only from shared structural features. Other terms, such as protein class, group, clan, and subfamily, have been coined over the years, but all suffer similar ambiguities of usage. A common usage is that superfamilies' (structural homology) contain families (sequence homology), which contain subfamilies. Hence, a superfamily, such as the PA clan of proteases, has far lower sequence conservation than one of the families it contains, the C04 family. an exact definition is unlikely to be agreed upon and to it is up to the reader to discern exactly how these terms are being used in a particular context.
Protein domains and motifs
The concept of protein family was conceived at a time when very few protein structures or sequences were known; at that time, primarily small, single-domain proteins such as myoglobin, hemoglobin, and cytochrome c were structurally understood. Since that time, many proteins were found to comprise multiple independent structural and functional units or domains. Due to evolutionary shuffling, different domains in a protein have evolved independently. This has led, in recent years, to a focus on families of protein domains. A number of online resources are devoted to identifying and cataloging such domains.
Regions of each protein have differing functional constraints (features critical to the structure and function of the protein). For example, the active site of an enzyme requires certain amino-acid residues to be precisely oriented in three dimensions. A protein–protein binding interface, though, may consist of a large surface with constraints on the hydrophobicity or polarity of the amino-acid residues. Functionally constrained regions of proteins evolve more slowly than unconstrained regions such as surface loops, giving rise to discernible blocks of conserved sequence when the sequences of a protein family are compared (see multiple sequence alignment). These blocks are most commonly referred to as motifs, although many other terms are used (blocks, signatures, fingerprints, etc.). Again, many online resources are devoted to identifying and cataloging protein motifs.
Evolution of protein families
According to current consensus, protein families arise in two ways. First, the separation of a parent species into two genetically isolated descendent species allows a gene/protein to independently accumulate variations (mutations) in these two lineages. This results in a family of orthologous proteins, usually with conserved sequence motifs. Second, a gene duplication may create a second copy of a gene (termed a paralog). Because the original gene is still able to perform its function, the duplicated gene is free to diverge and may acquire new functions (by random mutation). Certain gene/protein families, especially in eukaryotes, undergo extreme expansions and contractions in the course of evolution, sometimes in concert with whole genome duplications. This expansion and contraction of protein families is one of the salient features of genome evolution, but its importance and ramifications are currently unclear.
Use and importance of protein families
As the total number of sequenced proteins increases and interest expands in proteome analysis, an effort is ongoing to organize proteins into families and to describe their component domains and motifs. Reliable identification of protein families is critical to phylogenetic analysis, functional annotation, and the exploration of diversity of protein function in a given phylogenetic branch. The Enzyme Function Initiative is using protein families and superfamilies as the basis for development of a sequence/structure-based strategy for large scale functional assignment of enzymes of unknown function. The algorithmic means for establishing protein families on a large scale are based on a notion of similarity. Most of the time, the only similarity with access to is sequence similarity.
Protein family resources
Many biological databases record examples of protein families and allow users to identify if newly identified proteins belong to a known family. Here are a few examples:
- Pfam - Protein families database of alignments and HMMs
- PROSITE - Database of protein domains, families and functional sites
- PIRSF - SuperFamily Classification System
- PASS2 - Protein Alignment as Structural Superfamilies v2 - PASS2@NCBS
- SUPERFAMILY - Library of HMMs representing superfamilies and database of (superfamily and family) annotations for all completely sequenced organisms
- SCOP and CATH - classifications of protein structures into superfamilies, families and domains
Similarly, many database-searching algorithms exist, for example:
- BLAST - DNA sequence similarity search
- BLASTp - Protein sequence similarity search
- OrthoFinder a fast, scalable and accurate method for clustering proteins into families (orthogroups)
See also
Protein families
External links
-
 Media related to Protein families at Wikimedia Commons
Media related to Protein families at Wikimedia Commons
| Activity | |
|---|---|
| Regulation | |
| Classification | |
| Kinetics | |
| Types |
|