Pseudogene
Pseudogenes are nonfunctional segments of DNA that resemble functional genes. Most arise as superfluous copies of functional genes, either directly by gene duplication or indirectly by reverse transcription of an mRNA transcript. Pseudogenes are usually identified when genome sequence analysis finds gene-like sequences that lack regulatory sequences needed for transcription or translation, or whose coding sequences are obviously defective due to frameshifts or premature stop codons. Pseudogenes are a type of junk DNA.
Most non-bacterial genomes contain many pseudogenes, often as many as functional genes. This is not surprising, since various biological processes are expected to accidentally create pseudogenes, and there are no specialized mechanisms to remove them from genomes. Eventually pseudogenes may be deleted from their genomes by chance DNA replication or DNA repair errors, or they may accumulate so many mutational changes that they are no longer recognizable as former genes. Analysis of these degeneration events helps clarify the effects of non-selective processes in genomes.
Pseudogene sequences may be transcribed into RNA at low levels, due to promoter elements inherited from the ancestral gene or arising by new mutations. Although most of these transcripts will have no more functional significance than chance transcripts from other parts of the genome, some have given rise to beneficial regulatory RNAs and new proteins.
Properties
Pseudogenes are usually characterized by a combination of similarity to a known gene and loss of some functionality. That is, although every pseudogene has a DNA sequence that is similar to some functional gene, they are usually unable to produce functional final protein products. Pseudogenes are sometimes difficult to identify and characterize in genomes, because the two requirements of similarity and loss of functionality are usually implied through sequence alignments rather than biologically proven.
- Homology is implied by sequence similarity between the DNA sequences of the pseudogene and a known gene. After aligning the two sequences, the percentage of identical base pairs is computed. A high sequence identity means that it is highly likely that these two sequences diverged from a common ancestral sequence (are homologous), and highly unlikely that these two sequences have evolved independently (see Convergent evolution).
- Nonfunctionality can manifest itself in many ways. Normally, a gene must go through several steps to a fully functional protein: Transcription, pre-mRNA processing, translation, and protein folding are all required parts of this process. If any of these steps fails, then the sequence may be considered nonfunctional. In high-throughput pseudogene identification, the most commonly identified disablements are premature stop codons and frameshifts, which almost universally prevent the translation of a functional protein product.
Pseudogenes for RNA genes are usually more difficult to discover as they do not need to be translated and thus do not have "reading frames".
Pseudogenes can complicate molecular genetic studies. For example, amplification of a gene by PCR may simultaneously amplify a pseudogene that shares similar sequences. This is known as PCR bias or amplification bias. Similarly, pseudogenes are sometimes annotated as genes in genome sequences.
Processed pseudogenes often pose a problem for gene prediction programs, often being misidentified as real genes or exons. It has been proposed that identification of processed pseudogenes can help improve the accuracy of gene prediction methods.
Recently 140 human pseudogenes have been shown to be translated. However, the function, if any, of the protein products is unknown.
Types and origin
There are four main types of pseudogenes, all with distinct mechanisms of origin and characteristic features. The classifications of pseudogenes are as follows:
Processed
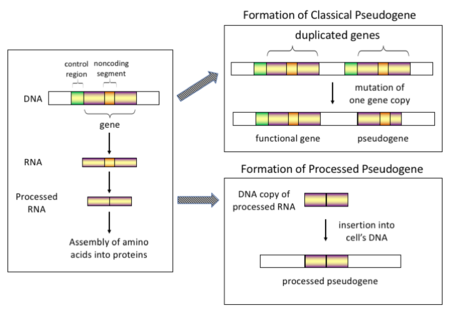
In higher eukaryotes, particularly mammals, retrotransposition is a fairly common event that has had a huge impact on the composition of the genome. For example, somewhere between 30 and 44% of the human genome consists of repetitive elements such as SINEs and LINEs (see retrotransposons). In the process of retrotransposition, a portion of the mRNA or hnRNA transcript of a gene is spontaneously reverse transcribed back into DNA and inserted into chromosomal DNA. Although retrotransposons usually create copies of themselves, it has been shown in an in vitro system that they can create retrotransposed copies of random genes, too. Once these pseudogenes are inserted back into the genome, they usually contain a poly-A tail, and usually have had their introns spliced out; these are both hallmark features of cDNAs. However, because they are derived from an RNA product, processed pseudogenes also lack the upstream promoters of normal genes; thus, they are considered "dead on arrival", becoming non-functional pseudogenes immediately upon the retrotransposition event. However, these insertions occasionally contribute exons to existing genes, usually via alternatively spliced transcripts. A further characteristic of processed pseudogenes is common truncation of the 5' end relative to the parent sequence, which is a result of the relatively non-processive retrotransposition mechanism that creates processed pseudogenes. Processed pseudogenes are continually being created in primates. Human populations, for example, have distinct sets of processed pseudogenes across its individuals.
Non-processed
Non-processed (or duplicated) pseudogenes. Gene duplication is another common and important process in the evolution of genomes. A copy of a functional gene may arise as a result of a gene duplication event caused by homologous recombination at, for example, repetitive sine sequences on misaligned chromosomes and subsequently acquire mutations that cause the copy to lose the original gene's function. Duplicated pseudogenes usually have all the same characteristics as genes, including an intact exon-intron structure and regulatory sequences. The loss of a duplicated gene's functionality usually has little effect on an organism's fitness, since an intact functional copy still exists. According to some evolutionary models, shared duplicated pseudogenes indicate the evolutionary relatedness of humans and the other primates. If pseudogenization is due to gene duplication, it usually occurs in the first few million years after the gene duplication, provided the gene has not been subjected to any selection pressure. Gene duplication generates functional redundancy and it is not normally advantageous to carry two identical genes. Mutations that disrupt either the structure or the function of either of the two genes are not deleterious and will not be removed through the selection process. As a result, the gene that has been mutated gradually becomes a pseudogene and will be either unexpressed or functionless. This kind of evolutionary fate is shown by population genetic modeling and also by genome analysis. According to evolutionary context, these pseudogenes will either be deleted or become so distinct from the parental genes so that they will no longer be identifiable. Relatively young pseudogenes can be recognized due to their sequence similarity.
Unitary pseudogenes
Various mutations (such as indels and nonsense mutations) can prevent a gene from being normally transcribed or translated, and thus the gene may become less- or non-functional or "deactivated". These are the same mechanisms by which non-processed genes become pseudogenes, but the difference in this case is that the gene was not duplicated before pseudogenization. Normally, such a pseudogene would be unlikely to become fixed in a population, but various population effects, such as genetic drift, a population bottleneck, or, in some cases, natural selection, can lead to fixation. The classic example of a unitary pseudogene is the gene that presumably coded the enzyme L-gulono-γ-lactone oxidase (GULO) in primates. In all mammals studied besides primates (except guinea pigs), GULO aids in the biosynthesis of ascorbic acid (vitamin C), but it exists as a disabled gene (GULOP) in humans and other primates. Another more recent example of a disabled gene links the deactivation of the caspase 12 gene (through a nonsense mutation) to positive selection in humans.
It has been shown that processed pseudogenes accumulate mutations faster than non-processed pseudogenes.
Pseudo-pseudogenes
The rapid proliferation of DNA sequencing technologies has led to the identification of many apparent pseudogenes using gene prediction techniques. Pseudogenes are often identified by the appearance of a premature stop codon in a predicted mRNA sequence, which would, in theory, prevent synthesis (translation) of the normal protein product of the original gene. There have been some reports of translational readthrough of such premature stop codons in mammals. As alluded to in the figure above, a small amount of the protein product of such readthrough may still be recognizable and function at some level. If so, the pseudogene can be subject to natural selection. That appears to have happened during the evolution of Drosophila species.
In 2016 it was reported that four predicted pseudogenes in multiple Drosophila species actually encode proteins with biologically important functions, "suggesting that such 'pseudo-pseudogenes' could represent a widespread phenomenon". For example, the functional protein (an olfactory receptor) is found only in neurons. This finding of tissue-specific biologically-functional genes that could have been classified as pseudogenes by in silico analysis complicates the analysis of sequence data. In the human genome, a number of examples have been identified that were originally classified as pseudogenes but later discovered to have a functional, although not necessarily protein-coding, role. As of 2012, it appeared that there are approximately 12,000–14,000 pseudogenes in the human genome. A 2016 proteogenomics analysis using mass spectrometry of peptides identified at least 19,262 human proteins produced from 16,271 genes or clusters of genes, with 8 new protein-coding genes identified that were previously considered pseudogenes.
Examples of pseudogene function
While the vast majority of pseudogenes have lost their function, some cases have emerged in which a pseudogene either re-gained its original or a similar function or evolved a new function. Examples include the following:
Drosophila glutamate receptor. The term "pseudo-pseudogene" was coined for the gene encoding the chemosensory ionotropic glutamate receptor Ir75a of Drosophila sechellia, which bears a premature termination codon (PTC) and was thus classified as a pseudogene. However, in vivo the D. sechellia Ir75a locus produces a functional receptor, owing to translational read-through of the PTC. Read-through is detected only in neurons and depends on the nucleotide sequence downstream of the PTC.
siRNAs. Some endogenous siRNAs appear to be derived from pseudogenes, and thus some pseudogenes play a role in regulating protein-coding transcripts, as reviewed. One of the many examples is psiPPM1K. Processing of RNAs transcribed from psiPPM1K yield siRNAs that can act to suppress the most common type of liver cancer, hepatocellular carcinoma. This and much other research has led to considerable excitement about the possibility of targeting pseudogenes with/as therapeutic agents
piRNAs. Some piRNAs are derived from pseudogenes located in piRNA clusters. Those piRNAs regulate genes via the piRNA pathway in mammalian testes and are crucial for limiting transposable element damage to the genome.
microRNAs. There are many reports of pseudogene transcripts acting as microRNA decoys. Perhaps the earliest definitive example of such a pseudogene involved in cancer is the pseudogene of BRAF. The BRAF gene is a proto-oncogene that, when mutated, is associated with many cancers. Normally, the amount of BRAF protein is kept under control in cells through the action of miRNA. In normal situations, the amount of RNA from BRAF and the pseudogene BRAFP1 compete for miRNA, but the balance of the 2 RNAs is such that cells grow normally. However, when BRAFP1 RNA expression is increased (either experimentally or by natural mutations), less miRNA is available to control the expression of BRAF, and the increased amount of BRAF protein causes cancer. This sort of competition for regulatory elements by RNAs that are endogenous to the genome has given rise to the term ceRNA.
PTEN. The PTEN gene is a known tumor suppressor gene. The PTEN pseudogene, PTENP1 is a processed pseudogene that is very similar in its genetic sequence to the wild-type gene. However, PTENP1 has a missense mutation which eliminates the codon for the initiating methionine and thus prevents translation of the normal PTEN protein. In spite of that, PTENP1 appears to play a role in oncogenesis. The 3' UTR of PTENP1 mRNA functions as a decoy of PTEN mRNA by targeting micro RNAs due to its similarity to the PTEN gene, and overexpression of the 3' UTR resulted in an increase of PTEN protein level. That is, overexpression of the PTENP1 3' UTR leads to increased regulation and suppression of cancerous tumors. The biology of this system is basically the inverse of the BRAF system described above.
Potogenes. Pseudogenes can, over evolutionary time scales, participate in gene conversion and other mutational events that may give rise to new or newly functional genes. This has led to the concept that pseudogenes could be viewed as potogenes: potential genes for evolutionary diversification.
Misidentified pseudogenes
Sometimes genes are thought to be pseudogenes, usually based on bioinformatic analysis, but then turn out to be functional genes. Examples include the Drosophila jingwei gene which encodes a functional alcohol dehydrogenase enzyme in vivo.
Another example is the human gene encoding phosphoglycerate mutase which was thought to be a pseudogene but which turned out to be a functional gene, now named PGAM4. Mutations in it cause infertility.
Bacterial pseudogenes
Pseudogenes are found in bacteria. Most are found in bacteria that are not free-living; that is, they are either symbionts or obligate intracellular parasites. Thus, they do not require many genes that are needed by free-living bacteria, such as gene associated with metabolism and DNA repair. However, there is not an order to which functional genes are lost first. For example, the oldest pseudogenes in Mycobacterium leprae are in RNA polymerases and the biosynthesis of secondary metabolites while the oldest ones in Shigella flexneri and Shigella typhi are in DNA replication, recombination, and repair.
Since most bacteria that carry pseudogenes are either symbionts or obligate intracellular parasites, genome size eventually reduces. An extreme example is the genome of Mycobacterium leprae, an obligate parasite and the causative agent of leprosy. It has been reported to have 1,133 pseudogenes which give rise to approximately 50% of its transcriptome. The effect of pseudogenes and genome reduction can be further seen when compared to Mycobacterium marinum, a pathogen from the same family. Mycobacteirum marinum has a larger genome compared to Mycobacterium leprae because it can survive outside the host; therefore, the genome must contain the genes needed to do so.
Although genome reduction focuses on what genes are not needed by getting rid of pseudogenes, selective pressures from the host can sway what is kept. In the case of a symbiont from the Verrucomicrobiota phylum, there are seven additional copies of the gene coding the mandelalide pathway. The host, species from Lissoclinum, use mandelalides as part of its defense mechanism.
The relationship between epistasis and the domino theory of gene loss was observed in Buchnera aphidicola. The domino theory suggests that if one gene of a cellular process becomes inactivated, then selection in other genes involved relaxes, leading to gene loss. When comparing Buchnera aphidicola and Escherichia coli, it was found that positive epistasis furthers gene loss while negative epistasis hinders it.
See also
- List of disabled human pseudogenes
- Molecular evolution
- Molecular paleontology
- Pseudogene (database)
- Retroposon
- Retrotransposon
Further reading
- Gerstein M, Zheng D (August 2006). "The real life of pseudogenes". Scientific American. 295 (2): 48–55. Bibcode:2006SciAm.295b..48G. doi:10.1038/scientificamerican0806-48. PMID 16866288.
- Torrents D, Suyama M, Zdobnov E, Bork P (December 2003). "A genome-wide survey of human pseudogenes". Genome Research. 13 (12): 2559–67. doi:10.1101/gr.1455503. PMC 403797. PMID 14656963.
- Bischof JM, Chiang AP, Scheetz TE, Stone EM, Casavant TL, Sheffield VC, Braun TA (June 2006). "Genome-wide identification of pseudogenes capable of disease-causing gene conversion". Human Mutation. 27 (6): 545–52. doi:10.1002/humu.20335. PMID 16671097. S2CID 20219423.
External links
- Pseudogene interaction database, miRNA-pseudogene and protein-pseudogene interaction maps database
- Yale University pseudogene database
- Hoppsigen database (homologous processed pseudogenes)
- RCPedia - Processed Pseudogene database
| Repeated sequence |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Transposon |
|
||||||||||
| Gene duplication | |||||||||||
| See also | |||||||||||