Ubiquitin-conjugating enzyme
| Ubiquitin—protein ligase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 6.3.2.19 | ||||||||
| CAS no. | 74812-49-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Ubiquitin-conjugating enzyme, E2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | UBQ-conjugat_E2 | ||||||||
| Pfam | PF00179 | ||||||||
| InterPro | IPR000608 | ||||||||
| SMART | SM00212 | ||||||||
| PROSITE | PDOC00163 | ||||||||
| Membranome | 241 | ||||||||
| |||||||||
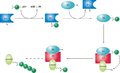
Ubiquitin-conjugating enzymes, also known as E2 enzymes and more rarely as ubiquitin-carrier enzymes, perform the second step in the ubiquitination reaction that targets a protein for degradation via the proteasome. The ubiquitination process covalently attaches ubiquitin, a short protein of 76 amino acids, to a lysine residue on the target protein. Once a protein has been tagged with one ubiquitin molecule, additional rounds of ubiquitination form a polyubiquitin chain that is recognized by the proteasome's 19S regulatory particle, triggering the ATP-dependent unfolding of the target protein that allows passage into the proteasome's 20S core particle, where proteases degrade the target into short peptide fragments for recycling by the cell.
Relationships
A ubiquitin-activating enzyme, or E1, first activates the ubiquitin by covalently attaching the molecule to its active site cysteine residue. The activated ubiquitin is then transferred to an E2 cysteine. Once conjugated to ubiquitin, the E2 molecule binds one of several ubiquitin ligases or E3s via a structurally conserved binding region. The E3 molecule is responsible for binding the target protein substrate and transferring the ubiquitin from the E2 cysteine to a lysine residue on the target protein.
A particular cell usually contains only a few types of E1 molecule, a greater diversity of E2s, and a very large variety of E3s. In humans, there are about 30 E2s which can bind with one of the 600+ E3s. The E3 molecules responsible for substrate identification and binding are thus the mechanisms of substrate specificity in proteasomal degradation. Each type of E2 can associate with many E3s.
E2s can also be used to study protein folding mechanisms. Since the ubiquitylation system is shared across all organisms, studies can use modified E2 proteins in order to understand the overall system for how all organisms process proteins. There are also some proteins which can act as both and E2 and an E3 containing domains which cover both E2 and E3 functionality.
Isozymes
The following human genes encode ubiquitin-conjugating enzymes:
- UBE2A
- UBE2B
- UBE2C
- UBE2D1, UBE2D2, UBE2D3, UBE2D4 (the latter putative)
- UBE2E1, UBE2E2, UBE2E3
- UBE2F (putative)
- UBE2G1, UBE2G2
- UBE2H
- UBE2I
- UBE2J1, UBE2J2
- UBE2K
- UBE2L3, UBE2L6; (UBE2L1, UBE2L2, UBE2L4 are pseudogenes)
- UBE2M
- UBE2N
- UBE2O
- UBE2Q1, UBE2Q2
- UBE2R1 (CDC34), UBE2R2
- UBE2S
- UBE2T (putative)
- UBE2U (putative)
- UBE2V1, UBE2V2
- UBE2W (putative)
- UBE2Z
- ATG3
- BIRC6
- UFC1
See also
External links
- Eukaryotic Linear Motif resource motif class MOD_SUMO
- Ubiquitin-Conjugating+Enzymes at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
|
Chaperones/ protein folding |
|
||||
|---|---|---|---|---|---|
| Protein targeting | |||||
|
Ubiquitin (ubiquitylation) |
|||||
|
Ubiquitin-like proteins (UBL) |
|
||||