
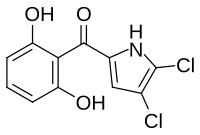
Pyoluteorin
 | |
| Identifiers | |
|---|---|
| |
| CAS Number | |
| PubChem CID | |
| ChemSpider | |
| UNII | |
| ChEBI | |
| ChEMBL | |
| CompTox Dashboard (EPA) | |
| Chemical and physical data | |
| Formula | C11H7Cl2NO3 |
| Molar mass | 272.08 g·mol−1 |
| 3D model (JSmol) | |
| |
| |
Pyoluteorin is a natural antibiotic that is biosynthesized from a hybrid nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) pathway. Pyoluteorin was first isolated in the 1950s from Pseudomonas aeruginosa strains T359 and IFO 3455 and was found to be toxic against oomycetes, bacteria, fungi, and against certain plants. Pyoluteorin is most notable for its toxicity against the oomycete Pythium ultimum, which is a plant pathogen that causes a global loss in agriculture. Currently, pyoluteorin derivatives are being studied as an Mcl-1 antagonist in order to target cancers that have elevated Mcl-1 levels.
Biosynthesis
Pyoluteorin is synthesized from an NRPS/PKS hybrid pathway. The resorcinol ring is derived from a type I PKS while the dichloropyrrolemoiety is derived from a type II NRPS. Pyoluteorin biosynthesis begins with the activation of L-proline to prolyl-AMP by the adenylation domain PltF. With prolyl-AMP still in the active site, the active form of the peptidyl carrier protein PltL binds to PltF. Then PltF catalyzes the aminoacylation of PltL by attaching L-proline to the thiol of the 4’phosphopantetheine arm of PltL. Next, the dehydrogenase PltE desaturates the prolyl moiety on PltL to create pyrrolyl-PltL. The halogenation domain PltA then dichlorinates the pyrrole moiety first at position 5 and then at position 4 in a FADH2 dependent manner. The dichloropyrroyl residue is then transferred to the type I PKS PltB and PltC, however, the mechanism of transfer is unknown. The addition of 3 malonyl-CoA monomers, cyclization, and release by the thioesterase PltG gives pyoluteorin.
