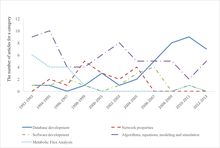
Systems biology
| Complex systems |
|---|
| Topics |
Systems biology is the computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological systems, using a holistic approach (holism instead of the more traditional reductionism) to biological research.
Particularly from the year 2000 onwards, the concept has been used widely in biology in a variety of contexts. The Human Genome Project is an example of applied systems thinking in biology which has led to new, collaborative ways of working on problems in the biological field of genetics. One of the aims of systems biology is to model and discover emergent properties, properties of cells, tissues and organisms functioning as a system whose theoretical description is only possible using techniques of systems biology. These typically involve metabolic networks or cell signaling networks.
Overview
Systems biology can be considered from a number of different aspects.
As a field of study, particularly, the study of the interactions between the components of biological systems, and how these interactions give rise to the function and behavior of that system (for example, the enzymes and metabolites in a metabolic pathway or the heart beats).
As a paradigm, systems biology is usually defined in antithesis to the so-called reductionist paradigm (biological organisation), although it is consistent with the scientific method. The distinction between the two paradigms is referred to in these quotations: "the reductionist approach has successfully identified most of the components and many of the interactions but, unfortunately, offers no convincing concepts or methods to understand how system properties emerge ... the pluralism of causes and effects in biological networks is better addressed by observing, through quantitative measures, multiple components simultaneously and by rigorous data integration with mathematical models." (Sauer et al.) "Systems biology ... is about putting together rather than taking apart, integration rather than reduction. It requires that we develop ways of thinking about integration that are as rigorous as our reductionist programmes, but different. ... It means changing our philosophy, in the full sense of the term." (Denis Noble)
As a series of operational protocols used for performing research, namely a cycle composed of theory, analytic or computational modelling to propose specific testable hypotheses about a biological system, experimental validation, and then using the newly acquired quantitative description of cells or cell processes to refine the computational model or theory. Since the objective is a model of the interactions in a system, the experimental techniques that most suit systems biology are those that are system-wide and attempt to be as complete as possible. Therefore, transcriptomics, metabolomics, proteomics and high-throughput techniques are used to collect quantitative data for the construction and validation of models.
As the application of dynamical systems theory to molecular biology. Indeed, the focus on the dynamics of the studied systems is the main conceptual difference between systems biology and bioinformatics.
As a socioscientific phenomenon defined by the strategy of pursuing integration of complex data about the interactions in biological systems from diverse experimental sources using interdisciplinary tools and personnel.
History
Systems biology was begun as a new field of science around 2000, when the Institute for Systems Biology was established in Seattle in an effort to lure "computational" type people who it was felt were not attracted to the academic settings of the university. The institute did not have a clear definition of what the field actually was: roughly bringing together people from diverse fields to use computers to holistically study biology in new ways. A Department of Systems Biology at Harvard Medical School was launched in 2003. In 2006 it was predicted that the buzz generated by the "very fashionable" new concept would cause all the major universities to need a systems biology department, thus that there would be careers available for graduates with a modicum of ability in computer programming and biology. In 2006 the National Science Foundation put forward a challenge to build a mathematical model of the whole cell. In 2012 the first whole-cell model of Mycoplasma genitalium was achieved by the Karr Laboratory at the Mount Sinai School of Medicine in New York. The whole-cell model is able to predict viability of M. genitalium cells in response to genetic mutations.
An earlier precursor of systems biology, as a distinct discipline, may have been by systems theorist Mihajlo Mesarovic in 1966 with an international symposium at the Case Institute of Technology in Cleveland, Ohio, titled Systems Theory and Biology. Mesarovic predicted that perhaps in the future there would be such as thing as "systems biology". Other early precursors that focused on the view that biology should be analyzed as a system, rather than a simple collection of parts, were Metabolic Control Analysis, developed by Henrik Kacser and Jim Burns later thoroughly revised, and Reinhart Heinrich and Tom Rapoport, and Biochemical Systems Theory developed by Michael Savageau
According to Robert Rosen in the 1960s, holistic biology had become passé by the early 20th century, as more empirical science dominated by molecular chemistry had become popular. Echoing him forty years later in 2006 Kling writes that the success of molecular biology throughout the 20th century had suppressed holistic computational methods. By 2011 the National Institutes of Health had made grant money available to support over ten systems biology centers in the United States, but by 2012 Hunter writes that systems biology had not lived up to the hype, having promised more than it achieved, which had caused it to become a somewhat minor field with few practical applications. Nonetheless, proponents hoped that it might once prove more useful in the future.
An important milestone in the development of systems biology has become the international project Physiome.
Associated disciplines
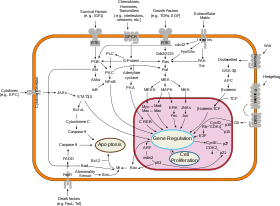
According to the interpretation of systems biology as using large data sets using interdisciplinary tools, a typical application is metabolomics, which is the complete set of all the metabolic products, metabolites, in the system at the organism, cell, or tissue level.
Items that may be a computer database include: phenomics, organismal variation in phenotype as it changes during its life span; genomics, organismal deoxyribonucleic acid (DNA) sequence, including intra-organismal cell specific variation. (i.e., telomere length variation); epigenomics/epigenetics, organismal and corresponding cell specific transcriptomic regulating factors not empirically coded in the genomic sequence. (i.e., DNA methylation, Histone acetylation and deacetylation, etc.); transcriptomics, organismal, tissue or whole cell gene expression measurements by DNA microarrays or serial analysis of gene expression; interferomics, organismal, tissue, or cell-level transcript correcting factors (i.e., RNA interference), proteomics, organismal, tissue, or cell level measurements of proteins and peptides via two-dimensional gel electrophoresis, mass spectrometry or multi-dimensional protein identification techniques (advanced HPLC systems coupled with mass spectrometry). Sub disciplines include phosphoproteomics, glycoproteomics and other methods to detect chemically modified proteins; glycomics, organismal, tissue, or cell-level measurements of carbohydrates; lipidomics, organismal, tissue, or cell level measurements of lipids.
The molecular interactions within the cell are also studied, this is called interactomics. A discipline in this field of study is protein-protein interactions, although interactomics includes the interactions of other molecules.Neuroelectrodynamics, where the computer's or a brain's computing function as a dynamic system is studied along with its (bio)physical mechanisms; and fluxomics, measurements of the rates of metabolic reactions in a biological system (cell, tissue, or organism).
In approaching a systems biology problem there are two main approaches. These are the top down and bottom up approach. The top down approach takes as much of the system into account as possible and relies largely on experimental results. The RNA-Seq technique is an example of an experimental top down approach. Conversely, the bottom up approach is used to create detailed models while also incorporating experimental data. An example of the bottom up approach is the use of circuit models to describe a simple gene network.
Various technologies utilized to capture dynamic changes in mRNA, proteins, and post-translational modifications. Mechanobiology, forces and physical properties at all scales, their interplay with other regulatory mechanisms;biosemiotics, analysis of the system of sign relations of an organism or other biosystems; Physiomics, a systematic study of physiome in biology.
Cancer systems biology is an example of the systems biology approach, which can be distinguished by the specific object of study (tumorigenesis and treatment of cancer). It works with the specific data (patient samples, high-throughput data with particular attention to characterizing cancer genome in patient tumour samples) and tools (immortalized cancer cell lines, mouse models of tumorigenesis, xenograft models, high-throughput sequencing methods, siRNA-based gene knocking down high-throughput screenings, computational modeling of the consequences of somatic mutations and genome instability). The long-term objective of the systems biology of cancer is ability to better diagnose cancer, classify it and better predict the outcome of a suggested treatment, which is a basis for personalized cancer medicine and virtual cancer patient in more distant prospective. Significant efforts in computational systems biology of cancer have been made in creating realistic multi-scale in silico models of various tumours.
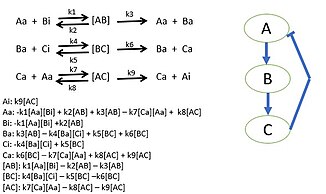
The systems biology approach often involves the development of mechanistic models, such as the reconstruction of dynamic systems from the quantitative properties of their elementary building blocks. For instance, a cellular network can be modelled mathematically using methods coming from chemical kinetics and control theory. Due to the large number of parameters, variables and constraints in cellular networks, numerical and computational techniques are often used (e.g., flux balance analysis).
Bioinformatics and data analysis
Other aspects of computer science, informatics, and statistics are also used in systems biology. These include new forms of computational models, such as the use of process calculi to model biological processes (notable approaches include stochastic π-calculus, BioAmbients, Beta Binders, BioPEPA, and Brane calculus) and constraint-based modeling; integration of information from the literature, using techniques of information extraction and text mining; development of online databases and repositories for sharing data and models, approaches to database integration and software interoperability via loose coupling of software, websites and databases, or commercial suits; network-based approaches for analyzing high dimensional genomic data sets. For example, weighted correlation network analysis is often used for identifying clusters (referred to as modules), modeling the relationship between clusters, calculating fuzzy measures of cluster (module) membership, identifying intramodular hubs, and for studying cluster preservation in other data sets; pathway-based methods for omics data analysis, e.g. approaches to identify and score pathways with differential activity of their gene, protein, or metabolite members. Much of the analysis of genomic data sets also include identifying correlations. Additionally, as much of the information comes from different fields, the development of syntactically and semantically sound ways of representing biological models is needed.
Creating biological models
Researchers begin by choosing a biological pathway and diagramming all of the protein interactions. After determining all of the interactions of the proteins, mass action kinetics is utilized to describe the speed of the reactions in the system. Mass action kinetics will provide differential equations to model the biological system as a mathematical model in which experiments can determine the parameter values to use in the differential equations. These parameter values will be the reaction rates of each proteins interaction in the system. This model determines the behavior of certain proteins in biological systems and bring new insight to the specific activities of individual proteins. Sometimes it is not possible to gather all reaction rates of a system. Unknown reaction rates are determined by simulating the model of known parameters and target behavior which provides possible parameter values.
The use of constraint-based reconstruction and analysis (COBRA) methods has become popular among systems biologists to simulate and predict the metabolic phenotypes, using genome-scale models. One of the methods is the flux balance analysis (FBA) approach, by which one can study the biochemical networks and analyze the flow of metabolites through a particular metabolic network, by maximizing the object of interest.
See also
- Biological computation
- BioSystems (journal)
- Computational biology
- Exposome
- Interactome
- List of omics topics in biology
- living systems
- Metabolic network modelling
- Modelling biological systems
- Molecular pathological epidemiology
- Network biology
- Network medicine
- Noogenesis
- Synthetic biology
- Systems biomedicine
- Systems immunology
- Systems medicine
- TIARA (database)
Further reading
- Klipp, Edda; Liebermeister, Wolfram; Wierling, Christoph; Kowald, Axel (2016). Systems Biology - A Textbook, 2nd edition. Wiley. ISBN 978-3-527-33636-4.
- Asfar S. Azmi, ed. (2012). Systems Biology in Cancer Research and Drug Discovery. ISBN 978-94-007-4819-4.
- Kitano, Hiroaki (15 October 2001). Foundations of Systems Biology. MIT Press. ISBN 978-0-262-11266-6.
- Werner, Eric (29 March 2007). "All systems go". Nature. 446 (7135): 493–494. Bibcode:2007Natur.446..493W. doi:10.1038/446493a. provides a comparative review of three books:
- Alon, Uri (7 July 2006). An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman & Hall. ISBN 978-1-58488-642-6.
- Kaneko, Kunihiko (15 September 2006). Life: An Introduction to Complex Systems Biology. Springer-Verlag. Bibcode:2006lics.book.....K. ISBN 978-3-540-32666-3.
- Palsson, Bernhard O. (16 January 2006). Systems Biology: Properties of Reconstructed Networks. Cambridge University Press. ISBN 978-0-521-85903-5.
- Werner Dubitzky; Olaf Wolkenhauer; Hiroki Yokota; Kwan-Hyun Cho, eds. (13 August 2013). Encyclopedia of Systems Biology. Springer-Verlag. ISBN 978-1-4419-9864-4.
External links
-
 Media related to Systems biology at Wikimedia Commons
Media related to Systems biology at Wikimedia Commons - Biological Systems in bio-physics-wiki
| |
| See also | |
| Genomics | |
|---|---|
| Bioinformatics | |
| Structural biology | |
| Research tools | |
| Organizations | |
|
System types |
|||||
|---|---|---|---|---|---|
| Concepts | |||||
| Theoretical fields |
|
||||
| Scientists |
|
||||
| Applications | |||||
| Organizations | |||||
| Authority control: National |
|---|