CRISPR gene editing
CRISPR gene editing (pronounced /ˈkrɪspər/ "crisper") is a genetic engineering technique in molecular biology by which the genomes of living organisms may be modified. It is based on a simplified version of the bacterial CRISPR-Cas9 antiviral defense system. By delivering the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) into a cell, the cell's genome can be cut at a desired location, allowing existing genes to be removed and/or new ones added in vivo.
The technique is considered highly significant in biotechnology and medicine as it enables editing genomes in vivo very precisely, cheaply, and easily. It can be used in the creation of new medicines, agricultural products, and genetically modified organisms, or as a means of controlling pathogens and pests. It also has possibilities in the treatment of inherited genetic diseases as well as diseases arising from somatic mutations such as cancer. However, its use in human germline genetic modification is highly controversial. The development of the technique earned Jennifer Doudna and Emmanuelle Charpentier the Nobel Prize in Chemistry in 2020. The third researcher group that shared the Kavli Prize for the same discovery, led by Virginijus Šikšnys, was not awarded the Nobel prize.
Working like genetic scissors, the Cas9 nuclease opens both strands of the targeted sequence of DNA to introduce the modification by one of two methods. Knock-in mutations, facilitated via homology directed repair (HDR), is the traditional pathway of targeted genomic editing approaches. This allows for the introduction of targeted DNA damage and repair. HDR employs the use of similar DNA sequences to drive the repair of the break via the incorporation of exogenous DNA to function as the repair template. This method relies on the periodic and isolated occurrence of DNA damage at the target site in order for the repair to commence. Knock-out mutations caused by CRISPR-Cas9 result from the repair of the double-stranded break by means of non-homologous end joining (NHEJ) or POLQ/ polymerase theta-mediated end-joining (TMEJ). These end-joining pathways can often result in random deletions or insertions at the repair site, which may disrupt or alter gene functionality. Therefore, genomic engineering by CRISPR-Cas9 gives researchers the ability to generate targeted random gene disruption. Because of this, the precision of genome editing is a great concern. Genomic editing leads to irreversible changes to the genome.
While genome editing in eukaryotic cells has been possible using various methods since the 1980s, the methods employed had proved to be inefficient and impractical to implement on a large scale. With the discovery of CRISPR and specifically the Cas9 nuclease molecule, efficient and highly selective editing is now a reality. Cas9 derived from the bacterial species Streptococcus pyogenes has facilitated targeted genomic modification in eukaryotic cells by allowing for a reliable method of creating a targeted break at a specific location as designated by the crRNA and tracrRNA guide strands. The ease with which researchers can insert Cas9 and template RNA in order to silence or cause point mutations at specific loci has proved invaluable to the quick and efficient mapping of genomic models and biological processes associated with various genes in a variety of eukaryotes. Newly engineered variants of the Cas9 nuclease have been developed that significantly reduce off-target activity.
CRISPR-Cas9 genome editing techniques have many potential applications, including in medicine and agriculture. The use of the CRISPR-Cas9-gRNA complex for genome editing was the AAAS's choice for Breakthrough of the Year in 2015. Many bioethical concerns have been raised about the prospect of using CRISPR for germline editing, especially in human embryos.
History
Other methods
In the early 2000s, German researchers began developing zinc finger nucleases (ZFNs), synthetic proteins whose DNA-binding domains enable them to create double-stranded breaks in DNA at specific points. ZFNs has a higher precision and the advantage of being smaller than Cas9, but ZFNs are not as commonly used as CRISPR-based methods. In 2010, synthetic nucleases called transcription activator-like effector nucleases (TALENs) provided an easier way to target a double-stranded break to a specific location on the DNA strand. Both zinc finger nucleases and TALENs require the design and creation of a custom protein for each targeted DNA sequence, which is a much more difficult and time-consuming process than that of designing guide RNAs. CRISPRs are much easier to design because the process requires synthesizing only a short RNA sequence, a procedure that is already widely used for many other molecular biology techniques (e.g. creating oligonucleotide primers).
Whereas methods such as RNA interference (RNAi) do not fully suppress gene function, CRISPR, ZFNs, and TALENs provide full irreversible gene knockout. CRISPR can also target several DNA sites simultaneously simply by introducing different gRNAs. In addition, the costs of employing CRISPR are relatively low.
Discovery
In 2012 Jennifer Doudna, Emmanuelle Charpentier, Martin Jínek, Krzysztof Chyliński, Ines Fonfara and Michael Hauer published their finding that CRISPR-Cas9 could be programmed with RNA to edit genomic DNA, now considered one of the most significant discoveries in the history of biology.
Patents and commercialization
As of November 2013, SAGE Labs (part of Horizon Discovery group) had exclusive rights from one of those companies to produce and sell genetically engineered rats and non-exclusive rights for mouse and rabbit models. By 2015, Thermo Fisher Scientific had licensed intellectual property from ToolGen to develop CRISPR reagent kits.
As of December 2014, patent rights to CRISPR were contested. Several companies formed to develop related drugs and research tools. As companies ramped up financing, doubts as to whether CRISPR could be quickly monetized were raised. In 2014, Feng Zhang of the Broad Institute of MIT and Harvard and nine others were awarded US patent number 8,697,359 over the use of CRISPR–Cas9 gene editing in eukaryotes. Although Charpentier and Doudna (referred to as CVC) were credited for the conception of CRISPR, the Broad Institute was the first to achieve a "reduction to practice" according to patent judges Sally Gardner Lane, James T. Moore and Deborah Katz.
The first set of patents was awarded to the Broad team in 2015, prompting attorneys for the CVC group to request the first interference proceeding. In February 2017 the US Patent Office ruled on a patent interference case brought by University of California with respect to patents issued to the Broad Institute, and found that the Broad patents, with claims covering the application of CRISPR-Cas9 in eukaryotic cells, were distinct from the inventions claimed by University of California.
Shortly after, University of California filed an appeal of this ruling. In 2019 the second interference dispute was opened. This was in response to patent applications made by CVC that required the appeals board to determine the original inventor of the technology. The USPTO ruled in March 2022 against UC, stating that the Broad Institute were first to file. The decision affected many of the licensing agreements for the CRISPR editing technology that was licensed from UC Berkeley. UC stated its intent to appeal the USPTO's ruling.
Recent events
In March 2017, the European Patent Office (EPO) announced its intention to allow claims for editing all types of cells to Max-Planck Institute in Berlin, University of California, and University of Vienna, and in August 2017, the EPO announced its intention to allow CRISPR claims in a patent application that MilliporeSigma had filed. As of August 2017 the patent situation in Europe was complex, with MilliporeSigma, ToolGen, Vilnius University, and Harvard contending for claims, along with University of California and Broad.
In July 2018, the ECJ ruled that gene editing for plants was a sub-category of GMO foods and therefore that the CRISPR technique would henceforth be regulated in the European Union by their rules and regulations for GMOs.
In February 2020, a US trial showed safe CRISPR gene editing on three cancer patients.
In October 2020, researchers Emmanuelle Charpentier and Jennifer Doudna were awarded the Nobel Prize in Chemistry for their work in this field. They made history as the first two women to share this award without a male contributor.
In June 2021, the first, small clinical trial of intravenous CRISPR gene editing in humans concludes with promising results.
In September 2021, the first CRISPR-edited food has gone on public sale in Japan. Tomatoes were genetically modified for around five times the normal amount of possibly calmingGABA. CRISPR was first applied in tomatoes in 2014.
In December 2021, it was reported that the first CRISPR-gene-edited marine animal/seafood and second set of CRISPR-edited food has gone on public sale in Japan: two fish of which one species grows to twice the size of natural specimens due to disruption of leptin, which controls appetite, and the other grows to 1.2 the natural average size with the same amount of food due to disabled myostatin, which inhibits muscle growth.
A 2022 study has found that knowing more about CRISPR tomatoes had a strong effect on the participants' preference. "Almost half of the 32 participants from Germany who are scientists demonstrated constant choices, while the majority showed increased willingness to buy CRISPR tomatoes, mostly non-scientists."
The UC Berkeley announced in May 2021 their intent to auction NFTs of both the patent for CRISPR gene editing as well as cancer immunotherapy. The university would in this case retain ownership of the patents, however. 85 % of funds gathered through the sale of the collection named The Fourth Pillar were to be used to finance research. It sold in June 2022 for 22 Ether, which was around US$54,000 at the time.
Genome engineering
CRISPR-Cas9 genome editing is carried out with a Type II CRISPR system. When utilized for genome editing, this system includes a ribonucleoprotein (RNP), consisting of Cas9, crRNA, and tracrRNA, along with an optional DNA repair template.
Major components
| Component | Function |
|---|---|
| crRNA | Contains the guide RNA that locates the correct segment of host DNA along with a region that binds to tracrRNA (generally in a hairpin loop form), forming an active complex. |
| tracrRNA | Binds to crRNA and forms an active complex. |
| sgRNA | Single-guide RNAs are a combined RNA consisting of a tracrRNA and at least one crRNA. |
| Cas9 (most commonly) | An enzyme whose active form is able to modify DNA. Many variants exist with different functions (i.e. single-strand nicking, double-strand breaking, DNA binding) due to each enzyme's DNA site recognition function. |
| Repair template | DNA molecule used as a template in the host cell's DNA repair process, allowing insertion of a specific DNA sequence into the host segment broken by Cas9. |
CRISPR-Cas9 often employs plasmids, that code for the RNP components, to transfect the target cells, or the RNP is assembled before addition to the cells via nucleofection. The main components of this plasmid are displayed in the image and listed in the table. The crRNA is uniquely designed for each application, as this is the sequence that Cas9 uses to identify and directly bind to specific sequences within the host cell's DNA. The crRNA must bind only where editing is desired. The repair template is also uniquely designed for each application, as it must complement to some degree the DNA sequences on either side of the cut and also contain whatever sequence is desired for insertion into the host genome.
Multiple crRNAs and the tracrRNA can be packaged together to form a single-guide RNA (sgRNA). This sgRNA can be included alongside the gene that codes for the Cas9 protein and made into a plasmid in order to be transfected into cells. Many online tools are available to aid in designing effective sgRNA sequences.
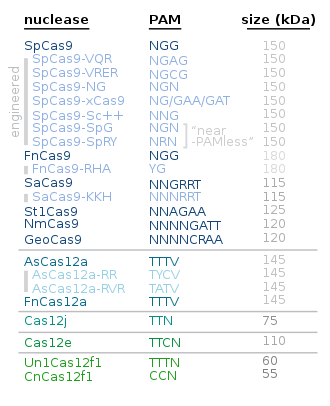
Alternatives to Cas9
Alternative proteins to Cas9 include the following:
| Protein | Main use / characteristics | Year/s |
|---|---|---|
| Cas12 | Cas12a is smaller and simpler than Cas9; Cas12b i.a. for plant genome engineering | |
| Cas13 | for RNA editing | |
| Cas3 | Creates a single-stranded wide gap | 2019 |
| CasMINI | About twice as compact as the more commonly used Cas9 and Cas12a. | 2021 |
| SuperFi-Cas9 | More accurate without a slow down in speed | 2022 |
| Cas7-11 | Better suited for therapeutic RNA editing than Cas13 | 2022 |
| Chromosome-templated DNA repair | Such a method is only applicable to organisms whose matching chromosome has the desired gene/s.
|
2022 |
Structure
CRISPR-Cas9 offers a high degree of fidelity and relatively simple construction. It depends on two factors for its specificity: the target sequence and the protospacer adjacent motif (PAM) sequence. The target sequence is 20 bases long as part of each CRISPR locus in the crRNA array. A typical crRNA array has multiple unique target sequences. Cas9 proteins select the correct location on the host's genome by utilizing the sequence to bond with base pairs on the host DNA. The sequence is not part of the Cas9 protein and as a result is customizable and can be independently synthesized.
The PAM sequence on the host genome is recognized by Cas9. Cas9 cannot be easily modified to recognize a different PAM sequence. However, this is ultimately not too limiting, as it is typically a very short and nonspecific sequence that occurs frequently at many places throughout the genome (e.g. the SpCas9 PAM sequence is 5'-NGG-3' and in the human genome occurs roughly every 8 to 12 base pairs).
Once these sequences have been assembled into a plasmid and transfected into cells, the Cas9 protein with the help of the crRNA finds the correct sequence in the host cell's DNA and – depending on the Cas9 variant – creates a single- or double-stranded break at the appropriate location in the DNA.
Properly spaced single-stranded breaks in the host DNA can trigger homology directed repair, which is less error-prone than the non-homologous end joining that typically follows a double-stranded break. Providing a DNA repair template allows for the insertion of a specific DNA sequence at an exact location within the genome. The repair template should extend 40 to 90 base pairs beyond the Cas9-induced DNA break. The goal is for the cell's native HDR process to utilize the provided repair template and thereby incorporate the new sequence into the genome. Once incorporated, this new sequence is now part of the cell's genetic material and passes into its daughter cells.
Delivery
Delivery of Cas9, sgRNA, and associated complexes into cells can occur via viral and non-viral systems. Electroporation of DNA, RNA, or ribonucleocomplexes is a common technique, though it can result in harmful effects on the target cells. Chemical transfection techniques utilizing lipids and peptides have also been used to introduce sgRNAs in complex with Cas9 into cells. Nanoparticle-based delivery has also been used for transfection. Types of cells that are more difficult to transfect (e.g., stem cells, neurons, and hematopoietic cells) require more efficient delivery systems, such as those based on lentivirus (LVs), adenovirus (AdV), and adeno-associated virus (AAV).
Efficiency of CRISPR-Cas9 has been found to greatly increase when various components of the system including the entire CRISPR/Cas9 structure to Cas9-gRNA complexes delivered in assembled form rather than using transgenics. This has found particular value in genetically modified crops for mass commercialization. Since the host's replication machinery is not needed to produce these proteins, the chance of the recognizing sequence of the sgRNA is almost none, decreasing the chance of off-target effects.
Controlled genome editing
Further improvements and variants of the CRISPR-Cas9 system have focused on introducing more control into its use. Specifically, the research aimed at improving this system includes improving its specificity, its efficiency, and the granularity of its editing power. Techniques can further be divided and classified by the component of the system they modify. These include using a different variants or novel creations of the Cas protein, using an altogether different effector protein, modifying the sgRNA, or using an algorithmic approach to identify existing optimal solutions.
Specificity is an important aspect to improve the CRISPR-Cas9 system because the off-target effects it generates have serious consequences for the genome of the cell and invokes caution for its use. Minimizing off-target effects is thus maximizing the safety of the system. Novel variations of Cas9 proteins that increase specificity include effector proteins with comparable efficiency and specificity to the original SpCas9 that are able to target the previously untargetable sequences and a variant that has virtually no off-target mutations. Research has also been conducted in engineering new Cas9 proteins, including some that partially replace RNA nucleotides in crRNA with DNA and a structure-guided Cas9 mutant generating procedure that all had reduced off-target effects. Iteratively truncated sgRNAs and highly stabilized gRNAs have been shown to also decrease off-target effects. Computational methods including machine learning have been used to predict the affinity of and create unique sequences for the system to maximize specificity for given targets.
Several variants of CRISPR-Cas9 allow gene activation or genome editing with an external trigger such as light or small molecules. These include photoactivatable CRISPR systems developed by fusing light-responsive protein partners with an activator domain and a dCas9 for gene activation, or by fusing similar light-responsive domains with two constructs of split-Cas9, or by incorporating caged unnatural amino acids into Cas9, or by modifying the guide RNAs with photocleavable complements for genome editing.
Methods to control genome editing with small molecules include an allosteric Cas9, with no detectable background editing, that will activate binding and cleavage upon the addition of 4-hydroxytamoxifen (4-HT), 4-HT responsive intein-linked Cas9, or a Cas9 that is 4-HT responsive when fused to four ERT2 domains. Intein-inducible split-Cas9 allows dimerization of Cas9 fragments and rapamycin-inducible split-Cas9 system developed by fusing two constructs of split-Cas9 with FRB and FKBP fragments. Other studies have been able to induce transcription of Cas9 with a small molecule, doxycycline. Small molecules can also be used to improve homology directed repair, often by inhibiting the non-homologous end joining pathway. A system with the Cpf1 effector protein was created that is induced by small molecules VE-822 and AZD-7762. These systems allow conditional control of CRISPR activity for improved precision, efficiency, and spatiotemporal control. Spatiotemporal control is a form of removing off-target effects—only certain cells or parts of the organism may need to be modified, and thus light or small molecules can be used as a way to conduct this. Efficiency of the CRISPR-Cas9 system is also greatly increased by proper delivery of the DNA instructions for creating the proteins and necessary reagents.
CRISPR also utilizes single base-pair editing proteins to create specific edits at one or two bases in the target sequence. CRISPR/Cas9 was fused with specific enzymes that initially could only change C to T and G to A mutations and their reverse. This was accomplished eventually without requiring any DNA cleavage. With the fusion of another enzyme, the base editing CRISPR-Cas9 system can also edit C to G and its reverse.
CRISPR screening
The clustered regularly interspaced short palindrome repeats (CRISPR)/Cas9 system is a gene-editing technology that can induce double-strand breaks (DSBs) anywhere guide ribonucleic acids (gRNA) can bind with the protospacer adjacent motif (PAM) sequence. Single-strand nicks can also be induced by Cas9 active-site mutants, also known as Cas9 nickases. By simply changing the sequence of gRNA, the Cas9-endonuclease can be delivered to a gene of interest and induce DSBs. The efficiency of Cas9-endonuclease and the ease by which genes can be targeted led to the development of CRISPR-knockout (KO) libraries both for mouse and human cells, which can cover either specific gene sets of interest or the whole-genome. CRISPR screening helps scientist to create a systematic and high-throughput genetic perturbation within live model organisms. This genetic perturbation is necessary for fully understanding gene function and epigenetic regulation. The advantage of pooled CRISPR libraries is that more genes can be targeted at once.
Knock-out libraries are created in a way to achieve equal representation and performance across all expressed gRNAs and carry an antibiotic or fluorescent selection marker that can be used to recover transduced cells. There are two plasmid systems in CRISPR/Cas9 libraries. First, is all in one plasmid, where sgRNA and Cas9 are produced simultaneously in a transfected cell. Second, is a two-vector system: sgRNA and Cas9 plasmids are delivered separately. It's important to deliver thousands of unique sgRNAs-containing vectors to a single vessel of cells by viral transduction at low multiplicity of infection (MOI, typically at 0.1-0.6), it prevents the probability that an individual cell clone will get more than one type of sgRNA otherwise it can lead to incorrect assignment of genotype to phenotype.
Once pooled library is prepared it is necessary to carry out a deep sequencing (NGS, next generation sequencing) of PCR-amplifed plasmid DNA in order to reveal abundance of sgRNAs. Cells of interest can be consequentially infected by the library and then selected according to the phenotype. There are 2 types of selection: negative and positive. By negative selection dead or slow growing cells are efficiently detected. It can identify survival-essential genes, which can be further serve as candidates for molecularly targeted drugs. On the other hand, positive selection gives a collection of growth-advantage acquired populations by random mutagenesis. After selection genomic DNA is collected and sequenced by NGS. Depletion or enrichment of sgRNAs is detected and compared to the original sgRNA library, annotated with the target gene that sgRNA corresponds to. Statistical analysis then identify genes that are significantly likely to be relevant to the phenotype of interest.
| Library | ID | Species | PI | Genes targeted | gRNAs per gene | Total gRNAs |
|---|---|---|---|---|---|---|
| Bassik Mouse CRISPR Knockout Library | 1000000121 — 1000000130 | Mouse | Bassik | Varies (~23,000 in total) | ~10 | Varies |
| Mouse Tumor Suppressor Gene CRISPR Knockout Library | 113584 EFS backbone
113585 TBG backbone |
Mouse | Chen | 56 | ~4 | 286 |
| Brie mouse genome-wide library | 73632 (1 plasmid)
73633 (2 plasmid) |
Mouse | Doench and Root | 19,674 | 4 | 78,637 |
| Bassik Human CRISPR Knockout Library | 101926 — 101934 | Human | Bassik | Varies (~20,500 in total) | ~10 | Varies |
| Brunello human genome-wide library | 73179 (1 plasmid)
73178 (2 plasmid) |
Human | Doench and Root | 19,114 | 4 | 76,441 |
| Mini-human AsCpf1-based Human Genome-wide Knockout Library | 130630 | Human | Draetta | 16,977 | 3-4 | 17,032 arrays |
Apart from knock-out there are also knock-down (CRISPRi) and activation (CRISPRa) libraries, which using the ability of proteolytically deactivated Cas9-fusion proteins (dCas9) to bind target DNA, which means that gene of interest is not cut but is over-expressed or repressed. It made CRISPR/Cas9 system even more interesting in gene editing. Inactive dCas9 protein modulate gene expression by targeting dCas9-repressors or activators toward promoter or transcriptional start sites of target genes. For repressing genes Cas9 can be fused to KRAB effector domain that makes complex with gRNA, whereas CRISPRa utilizes dCas9 fused to different transcriptional activation domains, which are further directed by gRNA to promoter regions to upregulate expression.
Applications
Disease models
Cas9 genomic modification has allowed for the quick and efficient generation of transgenic models within the field of genetics. Cas9 can be easily introduced into the target cells along with sgRNA via plasmid transfection in order to model the spread of diseases and the cell's response to and defense against infection. The ability of Cas9 to be introduced in vivo allows for the creation of more accurate models of gene function and mutation effects, all while avoiding the off-target mutations typically observed with older methods of genetic engineering.
The CRISPR and Cas9 revolution in genomic modeling does not extend only to mammals. Traditional genomic models such as Drosophila melanogaster, one of the first model organisms, have seen further refinement in their resolution with the use of Cas9. Cas9 uses cell-specific promoters allowing a controlled use of the Cas9. Cas9 is an accurate method of treating diseases due to the targeting of the Cas9 enzyme only affecting certain cell types. The cells undergoing the Cas9 therapy can also be removed and reintroduced to provide amplified effects of the therapy.
CRISPR-Cas9 can be used to edit the DNA of organisms in vivo and to eliminate individual genes or even entire chromosomes from an organism at any point in its development. Chromosomes that have been successfully deleted in vivo using CRISPR techniques include the Y chromosome and X chromosome of adult lab mice and human chromosomes 14 and 21, in embryonic stem cell lines and aneuploid mice respectively. This method might be useful for treating genetic disorders caused by abnormal numbers of chromosomes, such as Down syndrome and intersex disorders.
Successful in vivo genome editing using CRISPR-Cas9 has been shown in numerous model organisms, including Escherichia coli,Saccharomyces cerevisiae,Candida albicans, Methanosarcina acetivorans,Caenorhabditis elegans,Arabidopsis spp.,Danio rerio, and Mus musculus. Successes have been achieved in the study of basic biology, in the creation of disease models, and in the experimental treatment of disease models.
Concerns have been raised that off-target effects (editing of genes besides the ones intended) may confound the results of a CRISPR gene editing experiment (i.e. the observed phenotypic change may not be due to modifying the target gene, but some other gene). Modifications to CRISPR have been made to minimize the possibility of off-target effects. Orthogonal CRISPR experiments are often recommended to confirm the results of a gene editing experiment.
CRISPR simplifies the creation of genetically modified organisms for research which mimic disease or show what happens when a gene is knocked down or mutated. CRISPR may be used at the germline level to create organisms in which the targeted gene is changed everywhere (i.e. in all cells/tissues/organs of a multicellular organism), or it may be used in non-germline cells to create local changes that only affect certain cell populations within the organism.
CRISPR can be utilized to create human cellular models of disease. For instance, when applied to human pluripotent stem cells, CRISPR has been used to introduce targeted mutations in genes relevant to polycystic kidney disease (PKD) and focal segmental glomerulosclerosis (FSGS). These CRISPR-modified pluripotent stem cells were subsequently grown into human kidney organoids that exhibited disease-specific phenotypes. Kidney organoids from stem cells with PKD mutations formed large, translucent cyst structures from kidney tubules. The cysts were capable of reaching macroscopic dimensions, up to one centimeter in diameter. Kidney organoids with mutations in a gene linked to FSGS developed junctional defects between podocytes, the filtering cells affected in that disease. This was traced to the inability of podocytes to form microvilli between adjacent cells. Importantly, these disease phenotypes were absent in control organoids of identical genetic background, but lacking the CRISPR modifications.
A similar approach was taken to model long QT syndrome in cardiomyocytes derived from pluripotent stem cells. These CRISPR-generated cellular models, with isogenic controls, provide a new way to study human disease and test drugs.
Biomedicine
CRISPR-Cas technology has been proposed as a treatment for multiple human diseases, especially those with a genetic cause. Its ability to modify specific DNA sequences makes it a tool with potential to fix disease-causing mutations. Early research in animal models suggest that therapies based on CRISPR technology have potential to treat a wide range of diseases, including cancer,progeria, beta-thalassemia,sickle cell disease,hemophilia,cystic fibrosis,Duchenne's muscular dystrophy,Huntington's disease,transthyretin amyloidosis and heart disease. CRISPR has also been used to cure malaria in mosquitos, which could eliminate the vector and the disease in humans. CRISPR may also have applications in tissue engineering and regenerative medicine, such as by creating human blood vessels that lack expression of MHC class II proteins, which often cause transplant rejection.
In addition, clinical trials to cure beta thalassemia and sickle cell disease in human patients using CRISPR-Cas9 technology have shown promising results.
Nevertheless, there remains a few limitations of the technology's use in gene therapy: the relatively high frequency of off-target effect, the requirement for a PAM sequence near the target site, p53 mediated apoptosis by CRISPR-induced double-strand breaks and immunogenic toxicity due to the delivery system typically by virus.
Blindness
The CRISPR treatment for LCA10 (the most common variant of Leber Congenital Amaurosis which is the leading cause of inherited childhood blindness) modifies the patient's defective photoreceptor gene.
In March 2020, the first patient volunteer in this US-based study, sponsored by Editas Medicine, was given a low-dose of the treatment to test for safety.
In June 2021, enrollment began for a high-dose adult and pediatric cohort of 4 patient volunteers each. Dosing of the new cohorts is expected to be completed by July 2022. In November 2022, Editas reported that 20% of the patients treated had significant improvements, but also announced that the resulting target population was too small to support continued independent development.
Cancer
CRISPR has also found many applications in developing cell-based immunotherapies. The first clinical trial involving CRISPR started in 2016. It involved taking immune cells from people with lung cancer, using CRISPR to edit out the gene expressed PD-1, then administrating the altered cells back to the same person. 20 other trials were under way or nearly ready, mostly in China, as of 2017.
In 2016, the United States Food and Drug Administration (FDA) approved a clinical trial in which CRISPR would be used to alter T cells extracted from people with different kinds of cancer and then administer those engineered T cells back to the same people.
In November 2020, in mouse animal models, CRISPR was used effectively to treat glioblastoma (fast-growing brain tumor) and metastatic ovarian cancer, as those are two cancers with some of the worst best-case prognosis and are typically diagnosed during their later stages. The treatments have resulted in inhibited tumor growth, and increased survival by 80% for metastatic ovarian cancer and tumor cell apoptosis, inhibited tumor growth by 50%, and improved survival by 30% for glioblastoma.
In October 2021, CRISPR Therapeutics announced results from their ongoing US-based Phase 1 trial for an allogeneic T cell therapy. These cells are sourced from healthy donors and are edited to attack cancer cells and avoid being seen as a threat by the recipient's immune system, and then multiplied into huge batches which can be given to large numbers of recipients.
In December 2022, a 13-year British girl that had been diagnosed with incurable T-Cell Acute Lymphoblastic Leukaemia was cured by doctors at Great Ormond Street Hospital, in the first documented use of therapeutic gene editing for this purpose, after undergoing six months of an experimental treatment, where previous attempts of other treatments failed. The procedure included reprogramming a healthy T-Cell to destroy the cancerous T-Cells to first rid her of Leukaemia, and then rebuilding her immune system from scratch using healthy immune cells. The team used BASE editing and had previously treated a case of acute lymphoblastic leukaemia in 2015 using TALENs.
Diabetes
Type 1 Diabetes is an endocrine disorder which results from a lack of pancreatic beta cells to produce insulin, a vital compound in transporting blood sugar to cells for producing energy. Researchers have been trying to transplant healthy beta cells. CRISPR is used to edit the cells in order to reduce the chance the patient's body will reject the transplant.
In February 2022, a phase 1 trial was conducted in which one patient volunteer received treatment.
HIV/AIDS
Human immunodeficiency virus or HIV, is a virus that attacks the body's immune system. While effective treatments exist which can allow patients to live healthy lives, HIV is retroactive meaning that it embeds an inactive version of itself in the human genome. CRISPR can be used to selectively remove the virus from the genome by designing guide RNA to target the retroactive HIV genome. One issue with this approach is that it requires the removal of the HIV genome from almost all cells, which can be difficult to realistically achieve.
Infection
CRISPR-Cas-based "RNA-guided nucleases" can be used to target virulence factors, genes encoding antibiotic resistance, and other medically relevant sequences of interest. This technology thus represents a novel form of antimicrobial therapy and a strategy by which to manipulate bacterial populations. Recent studies suggest a correlation between the interfering of the CRISPR-Cas locus and acquisition of antibiotic resistance. This system provides protection of bacteria against invading foreign DNA, such as transposons, bacteriophages, and plasmids. This system was shown to be a strong selective pressure for the acquisition of antibiotic resistance and virulence factor in bacterial pathogens.
Therapies based on CRISPR–Cas3 gene editing technology delivered by engineered bacteriophages could be used to destroy targeted DNA in pathogens. Cas3 is more destructive than the better known Cas9.
Research suggests that CRISPR is an effective way to limit replication of multiple herpesviruses. It was able to eradicate viral DNA in the case of Epstein–Barr virus (EBV). Anti-herpesvirus CRISPRs have promising applications such as removing cancer-causing EBV from tumor cells, helping rid donated organs for immunocompromised patients of viral invaders, or preventing cold sore outbreaks and recurrent eye infections by blocking HSV-1 reactivation. As of August 2016, these were awaiting testing.
Initial results in the treatment and cure of HIV have been rather successful, in 2021 9 out of 23 humanized mice treated with a combination of anti-retrovirals and CRISPR/Cas-9 which lead to the virus becoming undetectable, even after the usual rebound period. None of the two treatments alone had such an effect. Clinical trials in humans are to start in 2022.
CRISPR may revive the concept of transplanting animal organs into people. Retroviruses present in animal genomes could harm transplant recipients. In 2015, a team eliminated 62 copies of a particular retroviral DNA sequence from the pig genome in a kidney epithelial cell. Researchers recently demonstrated the ability to birth live pig specimens after removing these retroviruses from their genome using CRISPR for the first time.
Genetic anthropology
CRISPR-Cas9 can be used in investigating and identifying the genetic differences of humans to other apes, especially of the brain. For example, by reintroducing archaic gene variants into brain organoids to show a major impact on neurodevelopment or by knockout of a gene in embryonic stem cells to identify a genetic regulator that via early cell shape transition drives evolutionary expansion of the human forebrain.
By technique
Knockdown/activation
Using "dead" versions of Cas9 (dCas9) eliminates CRISPR's DNA-cutting ability, while preserving its ability to target desirable sequences. Multiple groups added various regulatory factors to dCas9s, enabling them to turn almost any gene on or off or adjust its level of activity. Like RNAi, CRISPR interference (CRISPRi) turns off genes in a reversible fashion by targeting, but not cutting a site. The targeted site is methylated, epigenetically modifying the gene. This modification inhibits transcription. These precisely placed modifications may then be used to regulate the effects on gene expressions and DNA dynamics after the inhibition of certain genome sequences within DNA. Within the past few years, epigenetic marks in different human cells have been closely researched and certain patterns within the marks have been found to correlate with everything ranging from tumor growth to brain activity. Conversely, CRISPR-mediated activation (CRISPRa) promotes gene transcription. Cas9 is an effective way of targeting and silencing specific genes at the DNA level. In bacteria, the presence of Cas9 alone is enough to block transcription. For mammalian applications, a section of protein is added. Its guide RNA targets regulatory DNA sequences called promoters that immediately precede the target gene.
Cas9 was used to carry synthetic transcription factors that activated specific human genes. The technique achieved a strong effect by targeting multiple CRISPR constructs to slightly different locations on the gene's promoter.
RNA editing
In 2016, researchers demonstrated that CRISPR from an ordinary mouth bacterium could be used to edit RNA. The researchers searched databases containing hundreds of millions of genetic sequences for those that resembled CRISPR genes. They considered the fusobacterium Leptotrichia shahii. It had a group of genes that resembled CRISPR genes, but with important differences. When the researchers equipped other bacteria with these genes, which they called C2c2, they found that the organisms gained a novel defense. C2c2 has later been renamed to Cas13a to fit the standard nomenclature for Cas genes.
Many viruses encode their genetic information in RNA rather than DNA that they repurpose to make new viruses. HIV and poliovirus are such viruses. Bacteria with Cas13 make molecules that can dismember RNA, destroying the virus. Tailoring these genes opened any RNA molecule to editing.
CRISPR-Cas systems can also be employed for editing of micro-RNA and long-noncoding RNA genes in plants.
Therapeutic applications
Directing edits to correct mutated sequences was first proposed and demonstrated in 1995. This initial work used synthetic RNA antisense oligonucleotides complementary to a pre-mature stop codon mutation in a dystrophin sequence to activate A-to-I editing of the stop codon to a read through codon in a model xenopus cell system. While this also led to nearby inadvertent A-to-I transitions, A to I (read as G) transitions can correct all three stop codons, but cannot create a stop codon. Therefore, the changes led >25% correction of the targeted stop codon with read through to a downstream luciferase reporter sequence. Follow on work by Rosenthal achieved editing of mutated mRNA sequence in mammalian cell culture by directing an oligonucleotide linked to a cytidine deaminase to correct a mutated cystic fibrosis sequence. More recently, CRISPR-Cas13 fused to deaminases has been employed to direct mRNA editing.
In 2022, Cas7-11, better suited for therapeutic RNA editing than Cas13, was reported. It enables sufficiently targeted cuts and an early version of it was used for in vitro editing in 2021.Comparison to DNA editing
Gene drive
Gene drives may provide a powerful tool to restore balance of ecosystems by eliminating invasive species. Concerns regarding efficacy, unintended consequences in the target species as well as non-target species have been raised particularly in the potential for accidental release from laboratories into the wild. Scientists have proposed several safeguards for ensuring the containment of experimental gene drives including molecular, reproductive, and ecological. Many recommend that immunization and reversal drives be developed in tandem with gene drives in order to overwrite their effects if necessary. There remains consensus that long-term effects must be studied more thoroughly particularly in the potential for ecological disruption that cannot be corrected with reversal drives.
In vitro genetic depletion
Unenriched sequencing libraries often have abundant undesired sequences. Cas9 can specifically deplete the undesired sequences with double strand breakage with up to 99% efficiency and without significant off-target effects as seen with restriction enzymes. Treatment with Cas9 can deplete abundant rRNA while increasing pathogen sensitivity in RNA-seq libraries.
Epigenome editing
Epigenome editing or epigenome engineering is a type of genetic engineering in which the epigenome is modified at specific sites using engineered molecules targeted to those sites (as opposed to whole-genome modifications). Whereas gene editing involves changing the actual DNA sequence itself, epigenetic editing involves modifying and presenting DNA sequences to proteins and other DNA binding factors that influence DNA function. By "editing” epigenomic features in this manner, researchers can determine the exact biological role of an epigenetic modification at the site in question.
The engineered proteins used for epigenome editing are composed of a DNA binding domain that target specific sequences and an effector domain that modifies epigenomic features. Currently, three major groups of DNA binding proteins have been predominantly used for epigenome editing: Zinc finger proteins, Transcription Activator-Like Effectors (TALEs) and nuclease deficient Cas9 fusions (CRISPR).Applications
Targeted regulation of disease-related genes may enable novel therapies for many diseases, especially in cases where adequate gene therapies are not yet developed or are inappropriate. While transgenerational and population level consequences are not fully understood, it may become a major tool for applied functional genomics and personalized medicine. As with RNA editing, it does not involve genetic changes and their accompanying risks. One example of a potential functional use of epigenome editing was described in 2021: repressing Nav1.7 gene expression via CRISPR-dCas9 which showed therapeutic potential in three mouse models of chronic pain.
In 2022, research assessed its usefulness in reducing tau protein levels, regulating a protein involved in Huntington's disease, targeting an inherited form of obesity, and Dravet syndrome.CRISPR-directed integrases
Combination of CRISPR-Cas9 with integrases enabled a technique for large edits without problematic double-stranded breaks, as demonstrated with PASTE in 2022. The researchers reported it could be used to deliver genes as long as 36,000 DNA base pairs to several types of human cells and thereby potentially for treating diseases caused by a large number of mutations.
Prime editing
Prime editing (or base editing) is a CRISPR refinement to accurately insert or delete sections of DNA. The CRISPR edits are not always perfect and the cuts can end up in the wrong place. Both issues are a problem for using the technology in medicine. Prime editing does not cut the double-stranded DNA but instead uses the CRISPR targeting apparatus to shuttle an additional enzyme to a desired sequence, where it converts a single nucleotide into another. The new guide, called a pegRNA, contains an RNA template for a new DNA sequence to be added to the genome at the target location. That requires a second protein, attached to Cas9: a reverse transcriptase enzyme, which can make a new DNA strand from the RNA template and insert it at the nicked site. Those three independent pairing events each provide an opportunity to prevent off-target sequences, which significantly increases targeting flexibility and editing precision. Prime editing was developed by researchers at the Broad Institute of MIT and Harvard in Massachusetts. More work is needed to optimize the methods.
Society and culture
Human germline modification
As of March 2015, multiple groups had announced ongoing research with the intention of laying the foundations for applying CRISPR to human embryos for human germline engineering, including labs in the US, China, and the UK, as well as US biotechnology company OvaScience. Scientists, including a CRISPR co-discoverer, urged a worldwide moratorium on applying CRISPR to the human germline, especially for clinical use. They said "scientists should avoid even attempting, in lax jurisdictions, germline genome modification for clinical application in humans" until the full implications "are discussed among scientific and governmental organizations". These scientists support further low-level research on CRISPR and do not see CRISPR as developed enough for any clinical use in making heritable changes to humans.
In April 2015, Chinese scientists reported results of an attempt to alter the DNA of non-viable human embryos using CRISPR to correct a mutation that causes beta thalassemia, a lethal heritable disorder. The study had previously been rejected by both Nature and Science in part because of ethical concerns. The experiments resulted in successfully changing only some of the intended genes, and had off-target effects on other genes. The researchers stated that CRISPR is not ready for clinical application in reproductive medicine. In April 2016, Chinese scientists were reported to have made a second unsuccessful attempt to alter the DNA of non-viable human embryos using CRISPR – this time to alter the CCR5 gene to make the embryo resistant to HIV infection.
In December 2015, an International Summit on Human Gene Editing took place in Washington under the guidance of David Baltimore. Members of national scientific academies of the US, UK, and China discussed the ethics of germline modification. They agreed to support basic and clinical research under certain legal and ethical guidelines. A specific distinction was made between somatic cells, where the effects of edits are limited to a single individual, and germline cells, where genome changes can be inherited by descendants. Heritable modifications could have unintended and far-reaching consequences for human evolution, genetically (e.g. gene–environment interactions) and culturally (e.g. social Darwinism). Altering of gametocytes and embryos to generate heritable changes in humans was defined to be irresponsible. The group agreed to initiate an international forum to address such concerns and harmonize regulations across countries.
In February 2017, the United States National Academies of Sciences, Engineering, and Medicine (NASEM) Committee on Human Gene Editing published a report reviewing ethical, legal, and scientific concerns of genomic engineering technology. The conclusion of the report stated that heritable genome editing is impermissible now but could be justified for certain medical conditions; however, they did not justify the usage of CRISPR for enhancement.
In November 2018, Jiankui He announced that he had edited two human embryos to attempt to disable the gene for CCR5, which codes for a receptor that HIV uses to enter cells. He said that twin girls, Lulu and Nana, had been born a few weeks earlier. He said that the girls still carried functional copies of CCR5 along with disabled CCR5 (mosaicism) and were still vulnerable to HIV. The work was widely condemned as unethical, dangerous, and premature. An international group of scientists called for a global moratorium on genetically editing human embryos.
Policy barriers to genetic engineering
Policy regulations for the CRISPR-Cas9 system vary around the globe. In February 2016, British scientists were given permission by regulators to genetically modify human embryos by using CRISPR-Cas9 and related techniques. However, researchers were forbidden from implanting the embryos and the embryos were to be destroyed after seven days.
The US has an elaborate, interdepartmental regulatory system to evaluate new genetically modified foods and crops. For example, the Agriculture Risk Protection Act of 2000 gives the United States Department of Agriculture the authority to oversee the detection, control, eradication, suppression, prevention, or retardation of the spread of plant pests or noxious weeds to protect the agriculture, environment, and economy of the US. The act regulates any genetically modified organism that utilizes the genome of a predefined "plant pest" or any plant not previously categorized. In 2015, Yinong Yang successfully deactivated 16 specific genes in the white button mushroom to make them non-browning. Since he had not added any foreign-species (transgenic) DNA to his organism, the mushroom could not be regulated by the USDA under Section 340.2. Yang's white button mushroom was the first organism genetically modified with the CRISPR-Cas9 protein system to pass US regulation.
In 2016, the USDA sponsored a committee to consider future regulatory policy for upcoming genetic modification techniques. With the help of the US National Academies of Sciences, Engineering, and Medicine, special interests groups met on April 15 to contemplate the possible advancements in genetic engineering within the next five years and any new regulations that might be needed as a result. In 2017, the Food and Drug Administration proposed a rule that would classify genetic engineering modifications to animals as "animal drugs", subjecting them to strict regulation if offered for sale and reducing the ability for individuals and small businesses to make them profitable.
In China, where social conditions sharply contrast with those of the West, genetic diseases carry a heavy stigma. This leaves China with fewer policy barriers to the use of this technology.
Recognition
In 2012 and 2013, CRISPR was a runner-up in Science Magazine's Breakthrough of the Year award. In 2015, it was the winner of that award. CRISPR was named as one of MIT Technology Review's 10 breakthrough technologies in 2014 and 2016. In 2016, Jennifer Doudna and Emmanuelle Charpentier, along with Rudolph Barrangou, Philippe Horvath, and Feng Zhang won the Gairdner International award. In 2017, Doudna and Charpentier were awarded the Japan Prize in Tokyo, Japan for their revolutionary invention of CRISPR-Cas9. In 2016, Charpentier, Doudna, and Zhang won the Tang Prize in Biopharmaceutical Science. In 2020, Charpentier and Doudna were awarded the Nobel Prize in Chemistry, the first such prize for an all-female team, "for the development of a method for genome editing."
See also
- CRISPR/Cas Tools
- The CRISPR Journal
- Eugenics
- DRACO
- Zinc finger
- Gene knockout
- Genetics
- Glossary of genetics
- Human Nature (2019 documentary film)
- LEAPER gene editing
- Make People Better (2022 documentary)
- RNAi
- SiRNA
- Surveyor nuclease assay
- Synthetic biology
|
Science journal |
|
|---|---|
| Authority control: National |
|---|