
Zinc finger

A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (Xenopus laevis) transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells.Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in Drosophila. It often appears as a metal-binding domain in multi-domain proteins.
Proteins that contain zinc fingers (zinc finger proteins) are classified into several different structural families. Unlike many other clearly defined supersecondary structures such as Greek keys or β hairpins, there are a number of types of zinc fingers, each with a unique three-dimensional architecture. A particular zinc finger protein's class is determined by this three-dimensional structure, but it can also be recognized based on the primary structure of the protein or the identity of the ligands coordinating the zinc ion. In spite of the large variety of these proteins, however, the vast majority typically function as interaction modules that bind DNA, RNA, proteins, or other small, useful molecules, and variations in structure serve primarily to alter the binding specificity of a particular protein.
Since their original discovery and the elucidation of their structure, these interaction modules have proven ubiquitous in the biological world and may be found in 3% of the genes of the human genome. In addition, zinc fingers have become extremely useful in various therapeutic and research capacities. Engineering zinc fingers to have an affinity for a specific sequence is an area of active research, and zinc finger nucleases and zinc finger transcription factors are two of the most important applications of this to be realized to date.
History
Zinc fingers were first identified in a study of transcription in the African clawed frog, Xenopus laevis in the laboratory of Aaron Klug. A study of the transcription of a particular RNA sequence revealed that the binding strength of a small transcription factor (transcription factor IIIA; TFIIIA) was due to the presence of zinc-coordinating finger-like structures. Amino acid sequencing of TFIIIA revealed nine tandem sequences of 30 amino acids, including two invariant pairs of cysteine and histidine residues. Extended x-ray absorption fine structure confirmed the identity of the zinc ligands: two cysteines and two histidines. The DNA-binding loop formed by the coordination of these ligands by zinc were thought to resemble fingers, hence the name. This was followed soon thereafter by the discovery of the Krüppel factor in Drosophila by the Schuh team in 1986. More recent work in the characterization of proteins in various organisms has revealed the importance of zinc ions in polypeptide stabilization.
The crystal structures of zinc finger-DNA complexes solved in 1991 and 1993 revealed the canonical pattern of interactions of zinc fingers with DNA. The binding of zinc finger is found to be distinct from many other DNA-binding proteins that bind DNA through the 2-fold symmetry of the double helix, instead zinc fingers are linked linearly in tandem to bind nucleic acid sequences of varying lengths. Zinc fingers often bind to a sequence of DNA known as the GC box. The modular nature of the zinc finger motif allows for a large number of combinations of DNA and RNA sequences to be bound with high degree of affinity and specificity, and is therefore ideally suited for engineering protein that can be targeted to and bind specific DNA sequences. In 1994, it was shown that an artificially-constructed three-finger protein can block the expression of an oncogene in a mouse cell line. Zinc fingers fused to various other effector domains, some with therapeutic significance, have since been constructed.
Domain
Zinc finger (Znf) domains are relatively small protein motifs that contain multiple finger-like protrusions that make tandem contacts with their target molecule. Some of these domains bind zinc, but many do not, instead binding other metals such as iron, or no metal at all. For example, some family members form salt bridges to stabilise the finger-like folds. They were first identified as a DNA-binding motif in transcription factor TFIIIA from Xenopus laevis (African clawed frog), however they are now recognised to bind DNA, RNA, protein, and/or lipid substrates. Their binding properties depend on the amino acid sequence of the finger domains and on the linker between fingers, as well as on the higher-order structures and the number of fingers. Znf domains are often found in clusters, where fingers can have different binding specificities. Znf motifs occur in several unrelated protein superfamilies, varying in both sequence and structure. They display considerable versatility in binding modes, even between members of the same class (e.g., some bind DNA, others protein), suggesting that Znf motifs are stable scaffolds that have evolved specialised functions. For example, Znf-containing proteins function in gene transcription, translation, mRNA trafficking, cytoskeleton organization, epithelial development, cell adhesion, protein folding, chromatin remodeling, and zinc sensing, to name but a few. Zinc-binding motifs are stable structures, and they rarely undergo conformational changes upon binding their target.
Classes
Initially, the term zinc finger was used solely to describe DNA-binding motif found in Xenopus laevis; however, it is now used to refer to any number of structures related by their coordination of a zinc ion. In general, zinc fingers coordinate zinc ions with a combination of cysteine and histidine residues. Originally, the number and order of these residues was used to classify different types of zinc fingers ( e.g., Cys2His2, Cys4, and Cys6). More recently, a more systematic method has been used to classify zinc finger proteins instead. This method classifies zinc finger proteins into "fold groups" based on the overall shape of the protein backbone in the folded domain. The most common "fold groups" of zinc fingers are the Cys2His2-like (the "classic zinc finger"), treble clef, and zinc ribbon.
The following table shows the different structures and their key features:
| Fold Group | Representative structure | Ligand placement |
|---|---|---|
| Cys2His2 |  |
Two ligands from a knuckle and two more from the c terminus of a helix. |
| Gag knuckle |  |
Two ligands from a knuckle and two more from a short helix or loop. |
| Treble clef | Two ligands from a knuckle and two more from the N-terminus of a helix. | |
| Zinc ribbon | 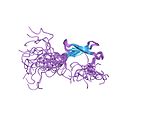 |
Two ligands each from two knuckles. |
| Zn2/Cys6 | 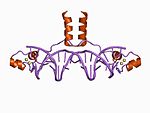 |
Two ligands from the N terminus of a helix and two more from a loop. |
| TAZ2 domain like | Two ligands from the termini of two helices. |
Cys2His2
| Zinc finger, C2H2 type | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | zf-C2H2 | ||||||||
| Pfam | PF00096 | ||||||||
| Pfam clan | CL0361 | ||||||||
| InterPro | IPR007087 | ||||||||
| PROSITE | PS00028 | ||||||||
| |||||||||
The Cys2His2-like fold group (C2H2) is by far the best-characterized class of zinc fingers, and is common in mammalian transcription factors. Such domains adopt a simple ββα fold and have the amino acid sequence motif:
- X2-Cys-X2,4-Cys-X12-His-X3,4,5-His
This class of zinc fingers can have a variety of functions such as binding RNA and mediating protein-protein interactions, but is best known for its role in sequence-specific DNA-binding proteins such as Zif268 (Egr1). In such proteins, individual zinc finger domains typically occur as tandem repeats with two, three, or more fingers comprising the DNA-binding domain of the protein. These tandem arrays can bind in the major groove of DNA and are typically spaced at 3-bp intervals. The α-helix of each domain (often called the "recognition helix") can make sequence-specific contacts to DNA bases; residues from a single recognition helix can contact four or more bases to yield an overlapping pattern of contacts with adjacent zinc fingers.
Gag-knuckle
| Zinc knuckle | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | zf-CCHC | ||||||||
| Pfam | PF00098 | ||||||||
| InterPro | IPR001878 | ||||||||
| SMART | SM00343 | ||||||||
| PROSITE | PS50158 | ||||||||
| |||||||||
This fold group is defined by two short β-strands connected by a turn (zinc knuckle) followed by a short helix or loop and resembles the classical Cys2His2 motif with a large portion of the helix and β-hairpin truncated.
The retroviral nucleocapsid (NC) protein from HIV and other related retroviruses are examples of proteins possessing these motifs. The gag-knuckle zinc finger in the HIV NC protein is the target of a class of drugs known as zinc finger inhibitors.
Treble-clef
The treble-clef motif consists of a β-hairpin at the N-terminus and an α-helix at the C-terminus that each contribute two ligands for zinc binding, although a loop and a second β-hairpin of varying length and conformation can be present between the N-terminal β-hairpin and the C-terminal α-helix. These fingers are present in a diverse group of proteins that frequently do not share sequence or functional similarity with each other. The best-characterized proteins containing treble-clef zinc fingers are the nuclear hormone receptors.
Zinc ribbon
| TFIIB zinc-binding | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | TF_Zn_Ribbon | ||||||||
| Pfam | PF08271 | ||||||||
| Pfam clan | Zn_Beta_Ribbon | ||||||||
| InterPro | IPR013137 | ||||||||
| PROSITE | PS51134 | ||||||||
| |||||||||
The zinc ribbon fold is characterised by two beta-hairpins forming two structurally similar zinc-binding sub-sites.
Zn2/Cys6
| Fungal Zn(2)-Cys(6) binuclear cluster domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Zn_clus | ||||||||
| Pfam | PF00172 | ||||||||
| InterPro | IPR001138 | ||||||||
| SMART | GAL4 | ||||||||
| PROSITE | PS00463 | ||||||||
| CDD | cd00067 | ||||||||
| |||||||||
The canonical members of this class contain a binuclear zinc cluster in which two zinc ions are bound by six cysteine residues. These zinc fingers can be found in several transcription factors including the yeast Gal4 protein.
| zf-C2HC | |||||||||
|---|---|---|---|---|---|---|---|---|---|
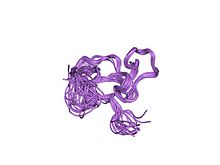 solution structure of a cchhc domain of neural zinc finger factor-1
| |||||||||
| Identifiers | |||||||||
| Symbol | zf-C2HC | ||||||||
| Pfam | PF01530 | ||||||||
| InterPro | IPR002515 | ||||||||
| |||||||||
| zf-C2HC5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | zf-C2HC5 | ||||||||
| Pfam | PF06221 | ||||||||
| InterPro | IPR009349 | ||||||||
| |||||||||
Miscellaneous
The zinc finger antiviral protein (ZAP) binds to the CpG site. It is used in mammals for antiviral defense.
Applications
Various protein engineering techniques can be used to alter the DNA-binding specificity of zinc fingers and tandem repeats of such engineered zinc fingers can be used to target desired genomic DNA sequences. Fusing a second protein domain such as a transcriptional activator or repressor to an array of engineered zinc fingers that bind near the promoter of a given gene can be used to alter the transcription of that gene. Fusions between engineered zinc finger arrays and protein domains that cleave or otherwise modify DNA can also be used to target those activities to desired genomic loci. The most common applications for engineered zinc finger arrays include zinc finger transcription factors and zinc finger nucleases, but other applications have also been described. Typical engineered zinc finger arrays have between 3 and 6 individual zinc finger motifs and bind target sites ranging from 9 basepairs to 18 basepairs in length. Arrays with 6 zinc finger motifs are particularly attractive because they bind a target site that is long enough to have a good chance of being unique in a mammalian genome.
Zinc finger nucleases
Engineered zinc finger arrays are often fused to a DNA cleavage domain (usually the cleavage domain of FokI) to generate zinc finger nucleases. Such zinc finger-FokI fusions have become useful reagents for manipulating genomes of many higher organisms including Drosophila melanogaster, Caenorhabditis elegans, tobacco, corn,zebrafish, various types of mammalian cells, and rats. Targeting a double-strand break to a desired genomic locus can be used to introduce frame-shift mutations into the coding sequence of a gene due to the error-prone nature of the non-homologous DNA repair pathway. If a homologous DNA "donor sequence" is also used then the genomic locus can be converted to a defined sequence via the homology directed repair pathway. An ongoing clinical trial is evaluating Zinc finger nucleases that disrupt the CCR5 gene in CD4+ human T-cells as a potential treatment for HIV/AIDS.
Methods of engineering zinc finger arrays
The majority of engineered zinc finger arrays are based on the zinc finger domain of the murine transcription factor Zif268, although some groups have used zinc finger arrays based on the human transcription factor SP1. Zif268 has three individual zinc finger motifs that collectively bind a 9 bp sequence with high affinity. The structure of this protein bound to DNA was solved in 1991 and stimulated a great deal of research into engineered zinc finger arrays. In 1994 and 1995, a number of groups used phage display to alter the specificity of a single zinc finger of Zif268. There are two main methods currently used to generate engineered zinc finger arrays, modular assembly, and a bacterial selection system, and there is some debate about which method is best suited for most applications.
The most straightforward method to generate new zinc finger arrays is to combine smaller zinc finger "modules" of known specificity. The structure of the zinc finger protein Zif268 bound to DNA described by Pavletich and Pabo in their 1991 publication has been key to much of this work and describes the concept of obtaining fingers for each of the 64 possible base pair triplets and then mixing and matching these fingers to design proteins with any desired sequence specificity. The most common modular assembly process involves combining separate zinc fingers that can each recognize a 3-basepair DNA sequence to generate 3-finger, 4-, 5-, or 6-finger arrays that recognize target sites ranging from 9 basepairs to 18 basepairs in length. Another method uses 2-finger modules to generate zinc finger arrays with up to six individual zinc fingers. The Barbas Laboratory of The Scripps Research Institute used phage display to develop and characterize zinc finger domains that recognize most DNA triplet sequences while another group isolated and characterized individual fingers from the human genome. A potential drawback with modular assembly in general is that specificities of individual zinc finger can overlap and can depend on the context of the surrounding zinc fingers and DNA. A recent study demonstrated that a high proportion of 3-finger zinc finger arrays generated by modular assembly fail to bind their intended target with sufficient affinity in a bacterial two-hybrid assay and fail to function as zinc finger nucleases, but the success rate was somewhat higher when sites of the form GNNGNNGNN were targeted.
A subsequent study used modular assembly to generate zinc finger nucleases with both 3-finger arrays and 4-finger arrays and observed a much higher success rate with 4-finger arrays. A variant of modular assembly that takes the context of neighboring fingers into account has also been reported and this method tends to yield proteins with improved performance relative to standard modular assembly.
Numerous selection methods have been used to generate zinc finger arrays capable of targeting desired sequences. Initial selection efforts utilized phage display to select proteins that bound a given DNA target from a large pool of partially randomized zinc finger arrays. This technique is difficult to use on more than a single zinc finger at a time, so a multi-step process that generated a completely optimized 3-finger array by adding and optimizing a single zinc finger at a time was developed. More recent efforts have utilized yeast one-hybrid systems, bacterial one-hybrid and two-hybrid systems, and mammalian cells. A promising new method to select novel 3-finger zinc finger arrays utilizes a bacterial two-hybrid system and has been dubbed "OPEN" by its creators. This system combines pre-selected pools of individual zinc fingers that were each selected to bind a given triplet and then utilizes a second round of selection to obtain 3-finger arrays capable of binding a desired 9-bp sequence. This system was developed by the Zinc Finger Consortium as an alternative to commercial sources of engineered zinc finger arrays. It is somewhat difficult to directly compare the binding properties of proteins generated with this method to proteins generated by modular assembly as the specificity profiles of proteins generated by the OPEN method have never been reported.
Examples
This entry represents the CysCysHisCys (C2HC) type zinc finger domain found in eukaryotes. Proteins containing these domains include:
- MYST family histone acetyltransferases
- Myelin transcription factor Myt1
- Suppressor of tumourigenicity protein 18 (ST18)
See also
External links
- C2H2 family Archived 2016-02-15 at the Wayback Machine at PlantTFDB: Plant Transcription Factor Database Archived 2016-04-22 at the Wayback Machine
- McDowall J. "Protein of the Month: Zinc Fingers". European Molecular Biology Laboratory - European Bioinformatics Institute (EMBL-EBI). Retrieved 2008-01-13.
- Goodsell DS. "Molecule of the Month: Zinc Fingers". Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB). Archived from the original on 2008-05-31. Retrieved 2008-01-13.
- The double helix between the zinc finger
- Zinc Finger Tools design and information site
- Human KZNF Gene Catalog Archived 2012-03-16 at the Wayback Machine
- Zinc finger C2H2-type domain Archived 2020-04-13 at the Wayback Machine in PROSITE
- Entry for zinc finger class C2H2 in the SMART database
- The Zinc Finger Consortium
- ZiFiT- Zinc Finger Design Tool
- Zinc Finger Consortium Materials from Addgene
- Predicting DNA-binding Specificities for C2H2 Zinc Finger Proteins
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
