
Mimiviridae
| Mimiviridae | |
|---|---|
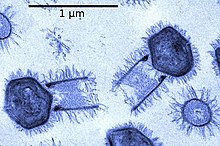
| |
| Tupanvirus | |
|
Virus classification | |
| (unranked): | Virus |
| Realm: | Varidnaviria |
| Kingdom: | Bamfordvirae |
| Phylum: | Nucleocytoviricota |
| Class: | Megaviricetes |
| Order: | Imitervirales |
| Family: | Mimiviridae |
| Subfamilies and genera | |
| |
Mimiviridae is a family of viruses. Amoeba and other protists serve as natural hosts. The family is divided in up to 4 subfamilies. Viruses in this family belong to the nucleocytoplasmic large DNA virus clade (NCLDV), also referred to as giant viruses.
Mimiviridae is the sole recognized member of order Imitervirales. Phycodnaviridae and Pandoraviridae of Algavirales are sister groups of Mimiviridae in many phylogenetic analyses.
History
The first member of this family, Mimivirus, was discovered in 2003, and the first complete genome sequence was published in 2004. However, the mimivirus Cafeteria roenbergensis virus was isolated and partially characterized in 1995, although the host was misidentified at the time, and the virus was designated BV-PW1.
Taxonomy
Group: dsDNA
- Family: Mimiviridae
Family Mimiviridae is currently divided into three subfamilies.
- One subfamily (genus Mimivirus, proposed names: Megavirinae or Megamimivirinae) is divided into three "lineages":
- A — Mimivirus group: includes Acanthamoeba polyphaga Mimivirus, Hirudovirus, Mamavirus, Kroon virus, Lentille virus, Terra2, Niemeyer virus, Samba virus.
- B — Moumouvirus group: includes Moumouvirus, Saudi moumouvirus, Moumouvirus goulette, Monve virus (aka Moumouvirus monve), and Ochan virus.
- C — Courdo11 virus group: includes Mont1, Courdo7, Courdo11, Megavirus chilensis, LBA111, Powai lake megavirus and Terra1.
- The majority of Mimiviridae appear to belong to this subfamily (Mimiviruses).
- It is sometimes also referred to as Mimiviridae group I.
- The second subfamily (Cafeteriavirus or Mimiviridae group II) includes the Cafeteria roenbergensis virus (CroV).
- The Klosneuvirinae have been proposed as a third subfamily and are divided into four "lineages": Klosneuvirus, Indivirus, Catovirus and Hokovirus. They seem to be closely related to the Mimivirus subfamily rather than the Cafeteriavirus subfamily (and so might be summarized in Mimivirus group I as well). The first isolate from this group is Bodo saltans virus infecting the kinetoplastid Bodo saltans.
- Tupanvirus strains have been discussed to comprise a sister group of mimiviruses.
Furthermore, it has been proposed either to extend Mimiviridae by an additional tentative group III (subfamily Mesomimivirinae) or to classify this group as a sister family Mesomimiviridae instead, comprising legacy OLPG (Organic Lake Phycodna Group). This extension (or sister family) may consist of the following:
- Phaeocystis globosa virus (PgV, represented by PgV-16T strain) and Phaeocystis pouchetii virus (PpV, e. g. PpV 01)
- "Organic Lake Phycodnavirus" 1 and 2 (OLV1, OLV2, hosts of Organic Lake virophage)
- "Yellowstone Lake Mimivirus" aka "Yellowstone Lake Phycodnavirus" 4 (YSLGV4)
- Chrysochromulina ericina virus (CeV, e. g. CeV 01)
- Aureococcus anophagefferens virus ( AaV)
- Pyramimonas orientalis virus (PoV)
- Tetraselmis virus (TetV-1)
This group seems to be closely related to Mimiviridae rather than to Phycodnaviridae and therefore is sometimes referred to as a further subfamily candidate Mesomimivirinae. Sometimes the extended family Mimiviridae is referred to as Megaviridae although this has not been recognized by ICTV; alternatively the extended group may be referred to just as Mimiviridae.
With recognition of new order Imitervirales by the ICTV in March 2020 there is no longer need to extend the Mimiviridae family to comprise a group of viruses of the observed high diversity. Instead, the extension (or at least its main clade) may be referred to as a sister family Mesomimiviridae.
Although only a couple of members of this order have been described in detail it seems likely there are many more awaiting description and assignment Unassigned members include Aureococcus anophagefferens virus (AaV), CpV-BQ2 and Terra2.
Structure
Viruses in Mimiviridae have icosahedral and round geometries, with between T=972 and T=1141, or T=1200 symmetry. The diameter is around 400 nm, with a length of 125 nm. Genomes are linear and non-segmented, around 1200kb in length. The genome has 911 open reading frames.
| Genus | Structure | Symmetry | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|
| Mimivirus | Icosahedral | T=972-1141 or T=1200 (H=19 +/- 1, K=19 +/- 1) | Linear | Monopartite |
| Klosneuvirus | Icosahedral | |||
| Cafeteriavirus | Icosahedral | T=499 | Linear | Monopartite |
| Tupanvirus | Tailed |
Life cycle
Replication follows the DNA strand displacement model. DNA-templated transcription is the method of transcription. Amoeba serve as the natural host.
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Mimivirus | Amoeba | None | Unknown | Unknown | Unknown | Unknown | Passive diffusion |
| Klosneuvirus | microzooplankton | None | Unknown | Unknown | Unknown | Cytoplasm | Passive diffusion |
| Cafeteriavirus | microzooplankton | None | Unknown | Unknown | Unknown | Cytoplasm | Passive diffusion |
Molecular biology
Within the genome of Lentille virus integrated genome of a virophage (Sputnik 2) and a transpoviron—a mobile genetic element—have been reported. Transpovirons are linear DNA elements of about 7 kilobases that encompass six to eight protein coding genes, two of which are homologous to virophage genes. Broad spectrum of mimiviridae virophage allows its isolation using a mimivirus reporter.
Clinical
Mimiviruses have been associated with pneumonia but their significance is currently unknown. The only virus of this family isolated from a human to date is LBA 111. At the Pasteur Institute of Iran (Tehran), researchers identified mimivirus DNA in bronchoalveolar lavage (BAL) and sputum samples of a child patient, utilizing real-time PCR (2018). Analysis reported 99% homology of LBA111, lineage C of the Megavirus chilensis. With only a few reported cases previous to this finding, the legitimacy of the mimivirus as an emerging infectious disease in humans remains controversial.
Mimivirus has also been implicated in rheumatoid arthritis.
See also
External links
