Rev (HIV)
| REV protein bound to RRE mRNA | |||||||||
|---|---|---|---|---|---|---|---|---|---|
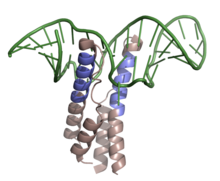 Shown above is the RNP complex formed by HIV-1 REV binding to the RRE upon mRNA of the env gene. Highlighted in slate is the ARM of the RNA binding domain, while colored in green is the mRNA with secondary stem-loop-like structure (PDB 4PMI).
| |||||||||
| Identifiers | |||||||||
| Symbol | REV | ||||||||
| Pfam | PF00424 | ||||||||
| InterPro | IPR000625 | ||||||||
| SCOP2 | 484d / SCOPe / SUPFAM | ||||||||
| |||||||||
Rev is a transactivating protein that is essential to the regulation of HIV-1 (and other lentiviral) protein expression. A nuclear localization signal is encoded in the rev gene, which allows the Rev protein to be localized to the nucleus, where it is involved in the export of unspliced and incompletely spliced mRNAs. In the absence of Rev, mRNAs of the HIV-1 late (structural) genes are retained in the nucleus, preventing their translation.
History
A novel protein was found to be involved in the translation of gag and env mRNA. The unknown protein functioned by removing repression of regulatory sequences and was named Art (anti-repression transactivator). Later studies suggested that the protein was involved in regulation of the RNA splicing mechanism. Therefore, the name of the protein was modified from Art to Trs (transregulator of splicing). The most recent studies have shown that the protein has multiple functions in the regulation of HIV-1 proteins, and its name has been changed to Rev (regulator of expression of virion proteins), which more generally describes its function.
Structure
Rev is a 13-kDa protein that is composed of 116 amino acids. Rev's sequence contains two specific domains which contribute to its nuclear import and export. The protein typically performs its function as a tetramer.
Arginine-rich motif
The N-terminal region of Rev contains an arginine-rich sequence. The arginine-rich motif (ARM) is located between amino acids 38–49 of the rev gene and forms an alpha-helical secondary structure. The ARM is a highly specific sequence which allows for the multimerization of Rev proteins, prior to RNA binding. A single base substitution alters Rev's ability to form a tetramer. The arginine-rich domain of Rev interacts with the rev-binding element (RBE), which is part of the HIV Rev response element (RRE) located in an intron downstream of the env gene. The alpha-helical secondary structure specifically can be considered a helix-loop-helix motif, which allows the REV protein to stably bind to the RRE RNA to form the ribonucleoprotein complex. The domain also contains a nuclear localization signal.
Rev-activation domain (Nuclear export signal)
Rev's nuclear export signal is located in residues 71–82 of the C-terminal region and is leucine-rich. Binding of Rev to viral RNAs containing the RRE allows for mRNA export out of the nucleus and into the cytoplasm by a mechanism different than that of cellular mRNAs.
Function
HIV-1 regulatory proteins (including Rev) are translated from completely processed mRNA transcripts, while structural proteins are translated from incompletely spliced transcripts. Completely spliced transcripts are exported from the nucleus to the cytoplasm by the same mechanism as cellular mRNA. However, Rev is needed to export incompletely spliced mRNAs in order to produce the viral structural proteins.
Rev localization to the nucleus
The arginine-rich domain of the Rev protein, containing a nuclear localization signal (NLS), allows Rev to enter the nucleus. Entry requires binding between a Rev multimer, Ran-GDP, and importin-β (a nuclear transport factor). The Rev NLS is a highly similar sequence to that of the importin-β-binding site present within importin-α, which allows for the interaction between Rev and importin-β. The NLS overlaps with the sequence required for RNA-binding. This prevents the NLS from counteracting the export of RRE-containing mRNA transcripts.
Binding of Rev to the RRE
The rev response element (RRE) is a 240 base-pair sequence located in the second intron of the HIV-1 genome, immediately downstream of the env gene. The RRE remains functional if translocated, but needs to remain in the same orientation (cannot be inverted). The RRE is retained by incompletely processed mRNA transcripts. The secondary structure of the RRE creates eight stem-loops. Rev initially binds to the purine-rich stem-loop IIB, then binds to a secondary site in stem-loop I.
Within this purine-rich stem-loop, IIB, are non-canonical base pairs that form as a result of the mRNA stem loop-secondary structure. These base pairs include guanine-adenine (nucleotides 47 and 73, respectively) and guanine-guanine (nucleotides 48–71, respectively). The two base pairs are separated by a non-stacked and bulging uridine that points outwards, away from the ARM-RNA interactions.
The ARM contains residues R35 and R39 that make base-specific contacts with residues on the RRE mRNA, specifically to bases uracil 66, guanine 67, and guanine 70, respectively. On the opposite side of these bases, residues N40 and R44 make base-specific contacts with nucleotides uracil 45, guanine 46, guanine 47, and adenine 73. In addition to these stabilizing contacts, additional Arg residues within the ARM, as well as T34, make nonspecific contacts with bases on the mRNA.
The RRE sequence is cis-acting, and is necessary to achieve high levels of env mRNA in the cytoplasm. The RRE also facilitates multimerization of the Rev proteins, which is required for Rev binding and function. The Rev protein binds unspliced gag and pol transcripts and incompletely spliced env, vif, vpr and vpu transcripts at the RRE, facilitating export to the cytoplasm.
Genomic export from the nucleus
Rev is continuously shuttled between the cytoplasm and nucleus. The shuttling of Rev is regulated by its nuclear localization signal and its nuclear export signal. Once Rev is inside the nucleus, Ran-GDP is phosphorylated into Ran-GTP, causing the importation complex to disassemble. Upon disassembly, Rev's NES forms a new complex with CRM1 (exportin-1) and Ran-GTP at the RRE sequence within incompletely spliced transcripts. Following assembly of the complex, the intron-containing RNAs are exported from the nucleus into the cytoplasm. Once the pre-mRNAs are in the cytoplasm, Rev dissociates, revealing the NLS. Exposure of the NLS allows for Rev interaction with importin-β in order to shuttle Rev back to the nucleus.
Rev-directed export of viral RNAs is similar to the mechanism by which snRNAs and the 5s rRNAs are exported, as opposed to the mechanism for export of cellular mRNAs. Rev is able to facilitate export of pre-mRNA transcripts that would otherwise typically remain in the nucleus, suggesting that the Rev NES is dominant over nuclear retention.
Regulation of HIV gene expression
Rev acts post-transcriptionally to positively regulate the expression of structural genes and to negatively regulate the expression of regulatory genes. Rev positively regulates the expression of gag, pol, and env. Rev-mediated export from the nucleus increases cytoplasmic levels of the structural mRNAs (gag, pol, and env).Gag, pol and env expression is lower in the absence of Rev and higher in the presence of Rev. Rev negatively regulates the expression of the regulatory genes (rev and tat) by creating a negative feedback loop, which regulates Rev production. There is a decrease in Rev production when Rev protein levels are higher than is necessary for the given amount of HIV-1 genome encoded. Rev also decreases the quantity of completely spliced viral messages expressed by exporting pre-mRNA before it can be spliced. This results in decreased expression of the regulatory proteins, Rev and Tat. Since Rev is continuously shuttled between the nucleus and cytoplasm, small amounts of the protein are able to impact many mRNA transcripts. Maintenance of the proper balance between early and late viral gene quantities leads to an overall increase in virion production.
Transition from early to late phase HIV-1 genes
HIV-1 genes are expressed from either completely spliced RNA or from intron-containing RNA. The export of fully spliced mRNAs (early, regulatory genes) occurs in the same manner as the normal export of cellular mRNAs. On the other hand, unspliced and incompletely spliced mRNAs which code for the late, structural proteins are Rev-dependent. The Rev protein is expressed as an early gene from completely spliced transcripts, so the expression of late phase structural proteins cannot occur until an initial amount of Rev is produced.
Rev as a target for antiviral therapies
Since Rev is absolutely necessary for HIV-1 replication and it is expressed early on in infection, it has been suggested that Rev is a good target for antiviral therapies.
Leptomycin B (LMB) binds to CRM1 which prevents the formation of the complex required for export(CRM1/NES/RanGTP/RRE) and ultimately reduces the production of incompletely spliced RNAs. Therefore, structural proteins, which are necessary for virion assembly, are not produced.
It has been shown that various organic compounds have the ability to target the Rev/RRE interaction. Neomycin B, diphenylfuran cation, and proflavine are small molecules that can prevent Rev from binding to the RRE sequence. If Rev is incapable of binding to the RRE on the pre-mRNA, the RNA will not be exported to the cytoplasm, also resulting in lack of necessary structural proteins.
Other therapies target the Rev protein itself, since it is an essential component of HIV-1 infection. M10 is a mutated form of Rev and has a single amino acid substitution (Aspartic acid to Leucine). If delivered to cells, Rev M10 will compete with the wild-type Rev protein for the RRE binding site, and therefore decrease Rev's normal cellular functions.
Dihydrovaltrate was also identified as a Rev-export inhibitory congener.
| DNA |
|
||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNA |
|
||||||||||||||||||||||||||||||||||||||||
| RT |
|
||||||||||||||||||||||||||||||||||||||||



