
Streptococcus pneumoniae
| Streptococcus pneumoniae | |
|---|---|
| |
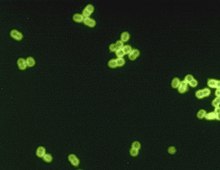
| S. pneumoniae in spinal fluid. FA stain (digitally colored). | |
|
Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Bacillota |
| Class: | Bacilli |
| Order: | Lactobacillales |
| Family: | Streptococcaceae |
| Genus: | Streptococcus |
| Species: |
S. pneumoniae
|
| Binomial name | |
|
Streptococcus pneumoniae (Klein 1884) Chester 1901
| |
Streptococcus pneumoniae, or pneumococcus, is a Gram-positive, spherical bacteria, alpha-hemolytic member of the genus Streptococcus. They are usually found in pairs (diplococci) and do not form spores and are non motile. As a significant human pathogenic bacterium S. pneumoniae was recognized as a major cause of pneumonia in the late 19th century, and is the subject of many humoral immunity studies.
Streptococcus pneumoniae resides asymptomatically in healthy carriers typically colonizing the respiratory tract, sinuses, and nasal cavity. However, in susceptible individuals with weaker immune systems, such as the elderly and young children, the bacterium may become pathogenic and spread to other locations to cause disease. It spreads by direct person-to-person contact via respiratory droplets and by auto inoculation in persons carrying the bacteria in their upper respiratory tracts. It can be a cause of neonatal infections.
Streptococcus pneumoniae is the main cause of community acquired pneumonia and meningitis in children and the elderly, and of sepsis in those infected with HIV. The organism also causes many types of pneumococcal infections other than pneumonia. These invasive pneumococcal diseases include bronchitis, rhinitis, acute sinusitis, otitis media, conjunctivitis, meningitis, sepsis, osteomyelitis, septic arthritis, endocarditis, peritonitis, pericarditis, cellulitis, and brain abscess.
Streptococcus pneumoniae can be differentiated from the viridans streptococci, some of which are also alpha-hemolytic, using an optochin test, as S. pneumoniae is optochin-sensitive. S. pneumoniae can also be distinguished based on its sensitivity to lysis by bile, the so-called "bile solubility test". The encapsulated, Gram-positive, coccoid bacteria have a distinctive morphology on Gram stain, lancet-shaped diplococci. They have a polysaccharide capsule that acts as a virulence factor for the organism; more than 100 different serotypes are known, and these types differ in virulence, prevalence, and extent of drug resistance.
History
In 1881, the organism, known later in 1886 as the pneumococcus for its role as a cause of pneumonia, was first isolated simultaneously and independently by the U.S. Army physician George Sternberg and the French chemist Louis Pasteur.
The organism was termed Diplococcus pneumoniae from 1920 because of its characteristic appearance in Gram-stained sputum. It was renamed Streptococcus pneumoniae in 1974 because it was very similar to streptococci.
Streptococcus pneumoniae played a central role in demonstrating that genetic material consists of DNA. In 1928, Frederick Griffith demonstrated transformation of life turning harmless pneumococcus into a lethal form by co-inoculating the live pneumococci into a mouse along with heat-killed virulent pneumococci. In 1944, Oswald Avery, Colin MacLeod, and Maclyn McCarty demonstrated that the transforming factor in Griffith's experiment was not protein, as was widely believed at the time, but DNA. Avery's work marked the birth of the molecular era of genetics.
Genetics
The genome of S. pneumoniae is a closed, circular DNA structure that contains between 2.0 and 2.1 million base pairs depending on the strain. It has a core set of 1553 genes, plus 154 genes in its virulome, which contribute to virulence and 176 genes that maintain a noninvasive phenotype. Genetic information can vary up to 10% between strains. The pneumococcal genome is known to contain a large and diverse repertoire of antimicrobial peptides, including 11 different lantibiotics.
Transformation
Natural bacterial transformation involves the transfer of DNA from one bacterium to another through the surrounding medium. Transformation is a complex developmental process requiring energy and is dependent on expression of numerous genes. In S. pneumoniae, at least 23 genes are required for transformation. For a bacterium to bind, take up, and recombine exogenous DNA into its chromosome, it must enter a special physiological state called competence. Competence in S. pneumoniae is induced by DNA-damaging agents such as mitomycin C, fluoroquinolone antibiotics (norfloxacin, levofloxacin and moxifloxacin), and topoisomerase inhibitors. Transformation protects S. pneumoniae against the bactericidal effect of mitomycin C. Michod et al. summarized evidence that induction of competence in S. pneumoniae is associated with increased resistance to oxidative stress and increased expression of the RecA protein, a key component of the recombinational repair machinery for removing DNA damages. On the basis of these findings, they suggested that transformation is an adaptation for repairing oxidative DNA damages. S. pneumoniae infection stimulates polymorphonuclear leukocytes (granulocytes) to produce an oxidative burst that is potentially lethal to the bacteria. The ability of S. pneumoniae to repair the oxidative DNA damages in its genome, caused by this host defense, likely contributes to this pathogen's virulence. Consistent with this premise, Li et al. reported that, among different highly transformable S. pneumoniae isolates, nasal colonization fitness and virulence (lung infectivity) depend on an intact competence system.
Infection
Streptococcus pneumoniae is part of the normal upper respiratory tract flora. As with many natural flora, it can become pathogenic under the right conditions, typically when the immune system of the host is suppressed. Invasins, such as pneumolysin, an antiphagocytic capsule, various adhesins, and immunogenic cell wall components are all major virulence factors. After S. pneumoniae colonizes the air sacs of the lungs, the body responds by stimulating the inflammatory response, causing plasma, blood, and white blood cells to fill the alveoli. This condition is called bacterial pneumonia.
Diseases and symptoms
Pneumonia is the most common of the S. pneumoniae diseases which include symptoms such as fever and chills, cough, rapid breathing, difficulty breathing, and chest pain. For the elderly, they may include confusion, low alertness, and the former listed symptoms to a lesser degree.
Pneumococcal meningitis is an infection of the tissue covering the brain and spinal cord. Symptoms include stiff neck, fever, headache, confusion, and photophobia.
Sepsis is caused by overwhelming response to an infection and leads to tissue damage, organ failure, and even death. The symptoms include confusion, shortness of breath, elevated heart rate, pain or discomfort, over-perspiration, fever, shivering, or feeling cold.
Vaccine
Due to the importance of disease caused by S. pneumoniae, several vaccines have been developed to protect against invasive infection. The World Health Organization recommends routine childhood pneumococcal vaccination; it is incorporated into the childhood immunization schedule in a number of countries including the United Kingdom, the United States, and South Africa.
Biotechnology
Components from S. pneumoniae have been harnessed for a range of applications in biotechnology. Through engineering of surface molecules from this bacterium, proteins can be irreversibly linked using the sortase enzyme or using the SnoopTag/SnoopCatcher reaction. Various glycoside hydrolases have also been cloned from S. pneumoniae to help analysis of cell glycosylation.
Interaction with Haemophilus influenzae
Historically, Haemophilus influenzae has been a significant cause of infection, and both H. influenzae and S. pneumoniae can be found in the human upper respiratory system. A study of competition in vitro revealed S. pneumoniae overpowered H. influenzae by attacking it with hydrogen peroxide. There is also evidence that S. pneumoniae uses hydrogen peroxide as a virulence factor. However, in a study adding both bacteria to the nasal cavity of a mouse within two weeks, only H. influenzae survives; further analysis showed that neutrophils exposed to dead H. influenzae were more aggressive in attacking S. pneumoniae.
Diagnosis
Diagnosis is generally made based on clinical suspicion along with a positive culture from a sample from virtually any place in the body. S. pneumoniae is, in general, optochin sensitive, although optochin resistance has been observed.
The recent advances in next-generation sequencing and comparative genomics have enabled the development of robust and reliable molecular methods for the detection and identification of S. pneumoniae. For instance, the Xisco gene was recently described as a biomarker for PCR-based detection of S. pneumoniae and differentiation from closely related species.
Atromentin and leucomelone possess antibacterial activity, inhibiting the enzyme enoyl-acyl carrier protein reductase, (essential for the biosynthesis of fatty acids) in S. pneumoniae.
Resistance
Resistant pneumococcal strains are called penicillin-resistant pneumococci (PRP), penicillin-resistant Streptococcus pneumoniae (PRSP),Streptococcus pneumoniae penicillin resistant (SPPR) or drug-resistant Strepotococcus pneumoniae (DRSP). In 2015, in the US, there were an estimated 30,000 cases, and in 30% of them the strains were resistant to one or more antibiotics.
See also
External links
- GAVI Alliance Archived 2014-08-20 at the Wayback Machine
- PneumoADIP
- PATH's Vaccine Resource Library pneumococcal resources
- Centers for Disease Control and Prevention (2012). "Ch. 16: Pneumococcal Disease". In Atkinson W; Wolfe S; Hamborsky J (eds.). Epidemiology and Prevention of Vaccine-Preventable Diseases (12th ed.). Washington DC: Public Health Foundation. pp. 233–248. Archived from the original on 2017-03-10.
- Type strain of Streptococcus pneumoniae at BacDive - the Bacterial Diversity Metadatabase Archived 2020-04-20 at the Wayback Machine
| Bacilli |
|
||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clostridia |
|
||||||||||||||||||||||||||||||||||
| Mollicutes |
|
||||||||||||||||||||||||||||||||||
