
Last universal common ancestor

The last universal common ancestor (LUCA) or universal most recent common ancestor (UMRCA) is the most recent population from which all organisms now living on Earth share common descent—the most recent common ancestor of all current life on Earth. This includes all cellular organisms, but not necessarilly viruses; the origins of viruses are unclear, however it is intriguing that they use the same genetic code as cellular organisms do: LUCA may have harboured a variety of viruses. The LUCA is not the first life on Earth; it may have lived among a diversity of other organisms whose descendants all died out. Rather LUCA is the most recent form from whom all surviving life on Earth is descended.
While no specific fossil evidence of the LUCA exists, the detailed biochemical similarity of all current life confirms its existence. Its characteristics can be inferred from shared features of modern genomes. These genes describe a complex life form with many co-adapted features, including transcription and translation mechanisms to convert information from DNA to RNA to proteins. The LUCA probably lived in the high-temperature water of deep sea vents near ocean-floor magma flows around 4 billion years ago.
Historical background

A phylogenetic tree directly portrays the idea of evolution by descent from a single ancestor. An early tree of life was sketched by Jean-Baptiste Lamarck in his Philosophie zoologique in 1809.Charles Darwin more famously proposed the theory of universal common descent through an evolutionary process in his book On the Origin of Species in 1859: "Therefore I should infer from analogy that probably all the organic beings which have ever lived on this earth have descended from some one primordial form, into which life was first breathed." The last sentence of the book begins with a restatement of the hypothesis:
- "There is grandeur in this view of life, with its several powers, having been originally breathed into a few forms or into one ..."
The term "last universal common ancestor" or "LUCA" was first used in the 1990s for such a primordial organism.
Inferring LUCA's features
An anaerobic thermophile
In 2016, Madeline C. Weiss and colleagues genetically analyzed 6.1 million protein-coding genes and 286,514 protein clusters from sequenced prokaryotic genomes representing many phylogenetic trees, and identified 355 protein clusters that were probably common to the LUCA. The results "depict LUCA as anaerobic, CO2-fixing, H2-dependent with a Wood–Ljungdahl pathway (the reductive acetyl-coenzyme A pathway), N2-fixing and thermophilic. LUCA's biochemistry was replete with FeS clusters and radical reaction mechanisms." The cofactors also reveal "dependence upon transition metals, flavins, S-adenosyl methionine, coenzyme A, ferredoxin, molybdopterin, corrins and selenium. Its genetic code required nucleoside modifications and S-adenosylmethionine-dependent methylations." The results are rather specific: they show that methanogenic clostridia was basal, near the root of the phylogenetic tree, in the 355 protein lineages examined, and that the LUCA may therefore have inhabited an anaerobic hydrothermal vent setting in a geochemically active environment rich in H2, CO2, and iron, where ocean water interacted with hot magma beneath the ocean floor.
While the gross anatomy of the LUCA can only be reconstructed with much uncertainty, its biochemical mechanisms can be described in some detail, based on the "universal" properties currently shared by all independently living organisms on Earth.

The LUCA certainly had genes and a genetic code. Its genetic material was most likely DNA, so that it lived after the RNA world. The DNA was kept double-stranded by an enzyme, DNA polymerase, which recognises the structure and directionality of DNA. The integrity of the DNA was maintained by a group of repair enzymes including DNA topoisomerase. If the genetic code was based on dual-stranded DNA, it was expressed by copying the information to single-stranded RNA. The RNA was produced by a DNA-dependent RNA polymerase using nucleotides similar to those of DNA. It had multiple DNA-binding proteins, such as histone-fold proteins. The genetic code was expressed into proteins. These were assembled from 20 free amino acids by translation of a messenger RNA via a mechanism of ribosomes, transfer RNAs, and a group of related proteins.
The LUCA was likely capable of sexual interaction in the sense that adaptive gene functions were present that promoted the transfer of DNA between individuals of the population to facilitate genetic recombination. Homologous gene products that promote genetic recombination are present in bacteria, archaea and eukaryota, such as the RecA protein in bacteria, the RadA protein in archaea, and the Rad51 and Dmc1 proteins in eukaryota.
As for the cell's gross structure, it contained a water-based cytoplasm effectively enclosed by a lipid bilayer membrane; it was capable of reproducing by cell division. It tended to exclude sodium and concentrate potassium by means of specific ion transporters (or ion pumps). The cell multiplied by duplicating all its contents followed by cellular division. The cell used chemiosmosis to produce energy. It also reduced CO2 and oxidized H2 (methanogenesis or acetogenesis) via acetyl-thioesters.
By phylogenetic bracketing, analysis of the presumed LUCA's offspring groups, LUCA appears to have been a small, single-celled organism. It likely had a ring-shaped coil of DNA floating freely within the cell. Morphologically, it would likely not have stood out within a mixed population of small modern-day bacteria. The originator of the three-domain system, Carl Woese, stated that in its genetic machinery, the LUCA would have been a "simpler, more rudimentary entity than the individual ancestors that spawned the three [domains] (and their descendants)".
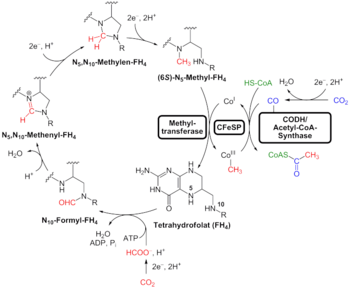
An alternative to the search for "universal" traits is to use genome analysis to identify phylogenetically ancient genes. This gives a picture of a LUCA that could live in a geochemically harsh environment and is like modern prokaryotes. Analysis of biochemical pathways implies the same sort of chemistry as does phylogenetic analysis. Weiss and colleagues write that "Experiments ... demonstrate that ... acetyl-CoA pathway [chemicals used in anaerobic respiration] formate, methanol, acetyl moieties, and even pyruvate arise spontaneously ... from CO2, native metals, and water", a combination present in hydrothermal vents.
Alternative interpretations
Some other researchers have challenged Weiss et al.'s 2016 conclusions. Sarah Berkemer and Shawn McGlynn argue that Weiss et al. undersampled the families of proteins, so that the phylogenetic trees were not complete and failed to describe the evolution of proteins correctly. A phylogenomic and geochemical analysis of a set of proteins that probably traced to the LUCA show that it had K+-dependent GTPases and the ionic composition and concentration of its intracellular fluid was seemingly high K+/Na+ ratio, NH+
4, Fe2+, CO2+, Ni2+, Mg2+, Mn2+, Zn2+, pyrophosphate, and PO3−
4 which would imply a terrestrial hot spring habitat. It possibly had a phosphate-based metabolism. Further, these proteins were unrelated to autotrophy (the ability of an organism to create its own organic matter), suggesting that the LUCA had a heterotrophic lifestyle (consuming organic matter) and that its growth was dependent on organic matter produced by the physical environment.Nick Lane argues that Na+/H+ antiporters could readily explain the low concentration of Na+ in the LUCA and its descendants.
The identification of thermophilic genes in the LUCA has also been criticized, as they may instead represent genes that evolved later in archaea or bacteria, then migrated between these via horizontal gene transfers, as in Woese's 1998 hypothesis. Further, the presence of the energy-handling enzymes CODH/acetyl-coenzyme A synthase in LUCA could be compatible not only with being an autotroph but also with life as a mixotroph or heterotroph. Weiss et al. 2018 reply that no enzyme defines a trophic lifestyle, and that heterotrophs evolved from autotrophs.
Age
Studies from 2000 to 2018 have suggested an increasingly ancient time for the LUCA. In 2000, estimates of the LUCA's age ranged from 3.5 to 3.8 billion years ago in the Paleoarchean, a few hundred million years before the earliest fossil evidence of life, for which candidates range in age from 3.48 to 4.28 billion years ago. This placed the LUCA shortly after the Late Heavy Asteroid Bombardment which was thought to have repeatedly sterilized Earth's surface. However, a 2018 study from the University of Bristol, applying a molecular clock model, found that the data (102 species, 29 common protein-coding genes, mostly ribosomal) is compatible only with LUCA as early as within 0.05 billion years of the maximum possible age given by the Earth-sterilizing Moon-forming event about 4.5 billion years ago.
Root of the tree of life
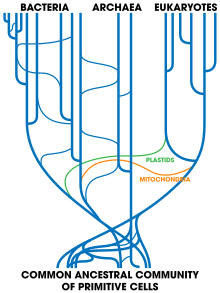
The most commonly accepted tree of life, based on several molecular studies, has its root between a monophyletic domain Bacteria and a clade formed by Archaea and Eukaryota. A small minority of studies place the root in the domain bacteria, in the phylum Bacillota, or state that the phylum Chloroflexota (formerly Chloroflexi) is basal to a clade with Archaea and Eukaryotes and the rest of bacteria (as proposed by Thomas Cavalier-Smith). Recent genomic analyses recover a two-domain system with the domains Archaea and Bacteria; in this view of the tree of life, Eukaryotes are derived from Archaea.
With the later gene pool of the LUCA's descendants, sharing a common framework of the AT/GC rule and the standard twenty amino acids, horizontal gene transfer would have become feasible and could have been common. In 1998, Carl Woese proposed that no individual organism could be considered a LUCA, and that the genetic heritage of all modern organisms derived through horizontal gene transfer among an ancient community of organisms. Other authors concur that there was a "complex collective genome" at the time of the LUCA, and that horizontal gene transfer was important in the evolution of later groups; Nicolas Glansdorff states that "Of course, LUCA was not living alone at that time (as the African Eve herself) but among many other organisms that have no descendants today." In 2010, based on "the vast array of molecular sequences now available from all domains of life," a "formal test" of universal common ancestry was published. The formal test favored the existence of a universal common ancestor over a wide class of alternative hypotheses that included horizontal gene transfer. Basic biochemical principles make it overwhelmingly likely that all organisms do have a single common ancestor. While the test overwhelmingly favored the existence of a single LUCA, this does not imply that the LUCA was ever alone: Instead, it was the only cell whose descendants survived beyond the Paleoarchean, outcompeting all others.
LUCA's viruses
Based on how viruses are currently distributed across the bacteria and archaea, the LUCA may well have been prey to multiple viruses, ancestral to those that now have those two domains as their hosts. Furthermore, extensive virus evolution seems to have preceded the LUCA, since the jelly-roll structure of capsid proteins is shared by RNA and DNA viruses across all three domains of life. LUCA's viruses were probably mainly dsDNA viruses in the groups called Duplodnaviria and Varidnaviria. Two other single-stranded DNA virus groups within the Monodnaviria, the Microviridae and the Tubulavirales, likely infected the last bacterial common ancestor. The last archaeal common ancestor was probably host to spindle-shaped viruses. All of these could well have affected the LUCA, in which case each must since have been lost in the host domain where it is no longer extant. By contrast, RNA viruses do not appear to have been important parasites of LUCA, even though straightforward thinking might have envisaged viruses as beginning with RNA viruses directly derived from an RNA world. Instead, by the time the LUCA lived, RNA viruses had probably already been out-competed by DNA viruses.
See also
- Abiogenesis – Natural process by which life arises from non-living matter
- Chemoton – Abstract model for the fundamental unit of life
- Darwinian threshold – Transition period during the evolution of the first cells
- Last eukaryotic common ancestor – the last common ancestor of animals, plants, and fungi
- Mitochondrial Eve – Matrilineal most recent common ancestor of all living humans
- Timeline of the evolutionary history of life – Major events during the development of life
- Urmetazoan – Hypothetical last common ancestor of all animals
- Y-chromosomal Adam – Patrilineal most recent common ancestor of all living humans
Further reading
- Lane, Nick (2016) [2015]. The Vital Question. London: Profile Books. ISBN 978-1781250372.
| History of research |
|
|---|---|
| Prebiotic synthesis | |
| Protocells | |
| Earliest organisms | |
| Research | |
|
Self-replicating organic structures
| |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellular life | |||||||||||
| Virus | |||||||||||
| Subviral agents |
|
||||||||||
|
Nucleic acid self-replication |
|
||||||||||
| Endosymbiosis | |||||||||||
| Abiogenesis | |||||||||||
| See also | |||||||||||
|
Life, non-cellular life, and comparable structures
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellular life |
|
||||||||||||||
| Non-cellular life |
|
||||||||||||||
| Comparable structures |
|
||||||||||||||

